In recent years, deep learning has revolutionized dermatology by automating skin cancer diagnosis with impressive accuracy. AI-powered systems like convolutional neural networks (CNNs) can now detect melanoma and other lesions with expert-level precision. However, alongside these advancements arises a critical vulnerability: adversarial attacks. These are subtle, often imperceptible image perturbations that can mislead even the most sophisticated AI models—potentially resulting in misdiagnosis.
Fortunately, a breakthrough solution is on the horizon. Researchers have proposed a novel, efficient, and highly effective defense method called the Multiscale Diffusive and Denoising Aggregation (MDDA) framework. This pioneering technique aims to reverse adversarial examples in skin cancer imaging and restore diagnostic accuracy—without the need for retraining or high computational cost.
What Are Adversarial Examples and Why Do They Matter in Dermatology?
Adversarial examples are intentionally modified images that appear normal to humans but are misleading to machine learning models. In medical imaging, such attacks can lead to grave consequences:
- False negatives: Cancerous lesions could be classified as benign.
- False positives: Harmless skin conditions could be mistaken for malignant ones.
- Security risks: Insurance fraud or erroneous AI-based clinical decisions.
These perturbations may be tiny, but their impact is massive—undermining trust in AI-driven medical tools.
Introducing MDDA: A Robust, Scalable Defense Framework
The MDDA framework is designed to “reverse-engineer” adversarial examples back to clean images. Unlike prior solutions that either require model access or expensive GPU resources, MDDA is:
- Attack-agnostic: Works against a variety of adversarial attacks.
- Training-free: No need to retrain the diagnostic model.
- Resource-efficient: Suitable for real-time use, even on portable devices.
It addresses one critical question in AI dermatology: Can we remove adversarial noise without losing diagnostic image quality?
How MDDA Works: Breaking Down the Technical Approach
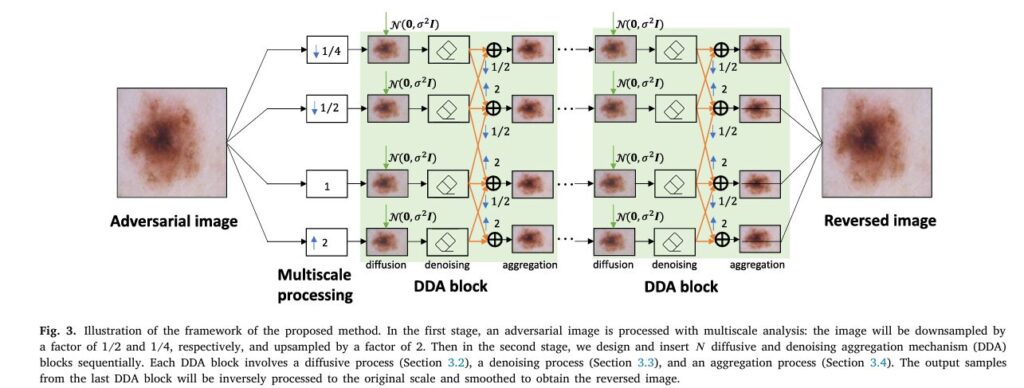
1. Multiscale Image Processing
MDDA begins by building an image pyramid. Each skin image is transformed across multiple scales—downsampled (¼x, ½x) and upsampled (2x)—to preserve both macro and micro-level features crucial for lesion classification. This multiscale approach enables better feature retention and structure analysis.
2. Diffusive Gaussian Noise Injection
Next, MDDA employs a “fight fire with fire” strategy. Controlled Gaussian noise is iteratively injected to drown out the adversarial patterns. The isotropic property of Gaussian noise ensures that injected signals cover a wide spectrum of directions—pushing the corrupted images back toward the clean data manifold.
3. Total Variation Denoising
After each noise injection, the image is denoised using a Total Variation (TV) regularization method, which is well-suited for preserving textures and edges. This process helps remove the residual noise while maintaining diagnostic integrity.
4. Cross-Scale Aggregation
To further refine the output, denoised images from adjacent scales are aggregated. This helps reinforce the diagnostic signal and minimize noise. The final image is reconstructed by averaging outputs from all scales, yielding a cleaned, adversary-free version ready for classification.
Experimental Results: MDDA Outperforms Existing Methods using Skin Cancer images
Dataset & Setup
- Dataset : ISIC 2019 (25,000+ dermoscopic images across 8 skin diseases).
- Models Tested : ResNet50 and MobileNetV2.
- Baseline Defenses : Bit Depth Reduction (BDR), Super-Resolution (SR), Neural Representation Purifier (NRP).
Performance Highlights
| DEFENSE METHODS | FGSM ACCURACY (%) | DIFGSM ACCURACY (%) | AUTOATTACK ACCURACY (%) |
|---|---|---|---|
| No Defense | 0.99 | 0.04 | 0 |
| BDR | 6.5 | 5.8 | 9.5 |
| SR | 6.2 | 7.4 | 22.8 |
| NRP | 30.6 | 38.5 | 39.1 |
| MDDA | 56.8 | 59.7 | 64.3 |
Key Takeaways
Balanced Clean Image Performance : Preserves 65.7% accuracy on unperturbed images, closely matching clinical standards.
Superior Accuracy : MDDA achieves 62.5% average accuracy under white-box attacks, outperforming NRP by 23%.
Cross-Architectural Defense : Maintains 57.7% accuracy when defending MobileNetV2 against ResNet50-crafted attacks.
Why MDDA Stands Out
| Feature | MDDA | Traditional Defenses |
|---|---|---|
| Model Agnostic | ✅ Yes | ❌ Often No |
| Requires Training | ❌ No | ✅ Yes |
| High Computational Demand | ❌ Low | ✅ High |
| Defense Across Attacks | ✅ Effective | ⚠️ Limited Scope |
| Suitable for Mobile Deployment | ✅ Yes | ❌ No |
MDDA’s smart use of diffusion-denoise cycles, combined with multiscale image processing, makes it a lightweight yet powerful adversarial defense framework—ideal for real-world deployment in dermatology clinics.
If you’re interested in Lung Cancer Diagnosis, you may also find this article helpful: LungCT-NET: Revolutionizing Lung Cancer Diagnosis with AI
Real-World Implications: Securing the Future of AI in Dermatology
As AI continues to assist dermatologists in early cancer detection, the need for reliable and secure models grows more urgent. The MDDA framework not only strengthens these models but does so in a practical and scalable manner. Its ability to reverse even complex adversarial attacks paves the way for:
- More trustworthy diagnostic tools
- Lower risk of insurance or diagnostic fraud
- Enhanced protection for AI-driven healthcare platforms
Conclusion: A New Era of Secure AI-Driven Healthcare
The MDDA framework represents a paradigm shift in defending against adversarial threats in medical imaging. Its combination of attack-agnostic resilience, computational efficiency, and cross-model compatibility positions it as a frontrunner in securing AI for healthcare. As adversarial tactics evolve, solutions like MDDA will be vital in safeguarding diagnostic integrity.
Call to Action
Are you working on AI security in healthcare? Join the conversation! Share your insights on adversarial defense mechanisms or explore collaboration opportunities with our research team. Together, we can build a safer, smarter future for medical AI.
Got questions about MDDA or adversarial attacks? Drop us a comment below!
FAQs
- What is an adversarial attack in medical imaging?
An adversarial attack is a technique where subtle, imperceptible modifications to medical images deceive AI models into making incorrect diagnoses. - How does MDDA improve skin cancer diagnosis?
MDDA reverses adversarial perturbations using multiscale noise diffusion and denoising, restoring image integrity and diagnostic accuracy. - Is MDDA compatible with all AI models?
Yes! MDDA is model-agnostic, functioning effectively across architectures like ResNet50 and MobileNetV2. - Can MDDA work on mobile devices?
Absolutely. Its resource-efficient design ensures smooth operation on edge devices, ideal for point-of-care applications. - Where can I learn more about the ISIC 2019 dataset?
Visit the ISIC Archive for access to the largest public dataset of dermoscopic skin cancer images.
Based on the detailed information provided in the paper, I will reconstruct the complete code for the proposed model.
import cv2
import os
import numpy as np
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torch.utils.data import Dataset, DataLoader
from torchvision import models, transforms
from torchvision.utils import save_image
import torchvision.transforms.functional as TF
from PIL import Image
import matplotlib.pyplot as plt
from sklearn.metrics import confusion_matrix, roc_curve, auc
from scipy.ndimage import gaussian_filter
from tqdm import tqdm
import cv2
import random
class MDDA:
"""
Multiscale Diffusive and Denoising Aggregation defense.
Implements the defense mechanism proposed in the paper.
"""
def __init__(self, sigma=0.125, num_dda_blocks=5, scales=(0.25, 0.5, 1.0, 2.0),
diffusion_steps=3, tv_weight=0.05):
"""
Args:
sigma: Variance of the Gaussian noise
num_dda_blocks: Number of DDA blocks
scales: Scales for multiscale processing
diffusion_steps: Number of diffusion steps
tv_weight: Weight for total variation regularization
"""
self.sigma = sigma
self.num_dda_blocks = num_dda_blocks
self.scales = scales
self.diffusion_steps = diffusion_steps
self.tv_weight = tv_weight
def transform(self, image, scale):
"""Apply scale transformation to an image while preserving device."""
if isinstance(image, torch.Tensor):
device = image.device
# Ensure we have 4D tensor for interpolation
original_dim = image.dim()
if image.dim() == 3:
image = image.unsqueeze(0)
elif image.dim() == 2:
image = image.unsqueeze(0).unsqueeze(0)
# Get current size
h, w = image.shape[2], image.shape[3]
new_h, new_w = int(h * scale), int(w * scale)
# Use F.interpolate instead of PIL to preserve device
if new_h > 0 and new_w > 0:
transformed = F.interpolate(
image,
size=(new_h, new_w),
mode='bilinear',
align_corners=False
)
else:
transformed = image
# Restore original dimensions
if original_dim == 3:
transformed = transformed.squeeze(0)
elif original_dim == 2:
transformed = transformed.squeeze(0).squeeze(0)
return transformed.to(device)
else:
# Numpy array
h, w = image.shape[:2]
new_size = (int(w * scale), int(h * scale))
return cv2.resize(image, new_size, interpolation=cv2.INTER_LINEAR)
def inverse_transform(self, image, scale):
"""Apply inverse scale transformation to an image."""
return self.transform(image, 1.0/scale)
def total_variation_loss(self, img):
"""Calculate total variation loss for denoising."""
if isinstance(img, torch.Tensor):
# Handle different tensor dimensions
if img.dim() == 4:
# 4D tensor: [batch, channels, height, width]
batch_size = img.size()[0]
h_x = img.size()[2]
w_x = img.size()[3]
count_h = (h_x - 1) * w_x
count_w = h_x * (w_x - 1)
h_tv = torch.pow(img[:, :, 1:, :] - img[:, :, :-1, :], 2).sum()
w_tv = torch.pow(img[:, :, :, 1:] - img[:, :, :, :-1], 2).sum()
return (h_tv + w_tv) / (count_h + count_w)
elif img.dim() == 3:
# 3D tensor: [channels, height, width]
h_x = img.size()[1]
w_x = img.size()[2]
count_h = (h_x - 1) * w_x
count_w = h_x * (w_x - 1)
h_tv = torch.pow(img[:, 1:, :] - img[:, :-1, :], 2).sum()
w_tv = torch.pow(img[:, :, 1:] - img[:, :, :-1], 2).sum()
return (h_tv + w_tv) / (count_h + count_w)
elif img.dim() == 2:
# 2D tensor: [height, width]
h_x = img.size()[0]
w_x = img.size()[1]
count_h = (h_x - 1) * w_x
count_w = h_x * (w_x - 1)
h_tv = torch.pow(img[1:, :] - img[:-1, :], 2).sum()
w_tv = torch.pow(img[:, 1:] - img[:, :-1], 2).sum()
return (h_tv + w_tv) / (count_h + count_w)
else:
raise ValueError(f"Unsupported tensor dimension: {img.dim()}. Expected 2D, 3D, or 4D tensor.")
else:
# Numpy version
h_tv = np.sum(np.power(img[1:, :] - img[:-1, :], 2))
w_tv = np.sum(np.power(img[:, 1:] - img[:, :-1], 2))
count_h = (img.shape[0] - 1) * img.shape[1]
count_w = img.shape[0] * (img.shape[1] - 1)
return (h_tv + w_tv) / (count_h + count_w)
def denoise(self, image, weight=0.1, num_iterations=50):
"""Denoise an image using total variation regularization."""
if isinstance(image, torch.Tensor):
device = image.device # Preserve device
# PyTorch implementation of TV denoising
original_shape = image.shape
# Ensure we have a 4D tensor for processing
if image.dim() == 3:
# Add batch dimension
image = image.unsqueeze(0)
added_batch_dim = True
elif image.dim() == 4:
added_batch_dim = False
else:
raise ValueError(f"Unsupported image dimension: {image.dim()}")
image = image.clone().detach()
image.requires_grad = True
optimizer = optim.Adamax([image], lr=0.01)
for _ in range(num_iterations):
# TV loss
tv_loss = self.total_variation_loss(image)
# Data fidelity term (MSE) - compare with original
loss = tv_loss * weight
optimizer.zero_grad()
loss.backward()
optimizer.step()
# Clamp values to [0, 1]
with torch.no_grad():
image.clamp_(0, 1)
result = image.detach()
# Remove batch dimension if we added it
if added_batch_dim:
result = result.squeeze(0)
return result.to(device) # Ensure result is on correct device
else:
# Numpy implementation (simplified)
denoised = cv2.bilateralFilter(image.astype(np.float32), 9, 75, 75)
return denoised
def diffuse(self, image, step):
"""Add isotropic Gaussian noise to an image."""
if isinstance(image, torch.Tensor):
device = image.device # Preserve device
# PyTorch implementation
noise = torch.randn_like(image, device=device) * (self.sigma / np.sqrt(self.diffusion_steps))
diffused = image + noise
return torch.clamp(diffused, 0, 1)
else:
# Numpy implementation
noise = np.random.normal(0, self.sigma / np.sqrt(self.diffusion_steps), image.shape)
diffused = image + noise
return np.clip(diffused, 0, 1)
def aggregate(self, images_dict):
"""Cleaner aggregate that normalizes all tensors to 4D and preserves device."""
# Get device from first tensor in dict
device = None
for v in images_dict.values():
if isinstance(v, torch.Tensor):
device = v.device
break
# Normalize all inputs to 4D tensors
normalized = {}
for k, v in images_dict.items():
if isinstance(v, torch.Tensor):
if v.dim() == 2: # (H, W)
v = v.unsqueeze(0).unsqueeze(0) # -> (1, 1, H, W)
elif v.dim() == 3: # (C, H, W)
v = v.unsqueeze(0) # -> (1, C, H, W)
elif v.dim() != 4:
raise ValueError(f"Unexpected tensor dimension at scale {k}: {v.dim()}")
normalized[k] = v.to(device) if device else v
else:
# Handle numpy arrays
if v.ndim == 2: # (H, W)
v = torch.from_numpy(v).unsqueeze(0).unsqueeze(0)
elif v.ndim == 3: # (H, W, C)
v = torch.from_numpy(v).permute(2, 0, 1).unsqueeze(0)
else:
raise ValueError(f"Unexpected array dimension at scale {k}: {v.ndim}")
normalized[k] = v.to(device) if device else v
result = {}
# Process each scale with 4D tensors
for k in normalized:
neighbors = []
for k_prime in normalized:
if k != k_prime:
scale_ratio = k_prime / k
transformed = self.transform(normalized[k_prime], scale_ratio)
# Ensure transformed is 4D and on correct device
if transformed.dim() < 4:
while transformed.dim() < 4:
transformed = transformed.unsqueeze(0)
if device:
transformed = transformed.to(device)
neighbors.append(transformed)
if neighbors:
aggregated = normalized[k].clone()
for neighbor in neighbors:
if neighbor.shape[2:] != aggregated.shape[2:]:
neighbor = F.interpolate(
neighbor,
size=(aggregated.shape[2], aggregated.shape[3]),
mode='bilinear',
align_corners=False
)
aggregated = aggregated + neighbor.to(aggregated.device)
aggregated = aggregated / (1 + len(neighbors))
result[k] = aggregated
else:
result[k] = normalized[k]
# Convert back to original dimensions
final_result = {}
for k in result:
output = result[k]
original = images_dict[k]
if isinstance(original, torch.Tensor):
# Match original dimensions
while output.dim() > original.dim():
output = output.squeeze(0)
else:
# Convert back to numpy if original was numpy
output = output.squeeze(0).permute(1, 2, 0).cpu().numpy()
final_result[k] = output
return final_result
def dda_block(self, images_dict):
"""Apply one DDA (Diffusion-Denoising-Aggregation) block."""
result = {}
# For each scale, apply diffusion and denoising
for k, image in images_dict.items():
current = image
# Apply diffusion-denoising for multiple steps
for t in range(self.diffusion_steps):
# Diffuse
current = self.diffuse(current, t)
# Denoise
current = self.denoise(current, weight=self.tv_weight)
result[k] = current
# Aggregate
result = self.aggregate(result)
return result
def defend(self, image):
"""
Apply MDDA defense to an adversarial image.
Args:
image: Input adversarial image (tensor or numpy array)
Returns:
defended_image: Image with adversarial noise removed
"""
is_tensor = isinstance(image, torch.Tensor)
original_shape = image.shape
# If it's a batch, process each image separately
if is_tensor and image.dim() == 4:
batch_size = image.shape[0]
defended_batch = []
for i in range(batch_size):
defended = self.defend_single(image[i])
defended_batch.append(defended)
return torch.stack(defended_batch)
else:
return self.defend_single(image)
def defend_single(self, image):
"""Apply MDDA defense to a single image with proper device handling."""
is_tensor = isinstance(image, torch.Tensor)
device = image.device if is_tensor else None
# Ensure image is 3D: [channels, height, width]
if is_tensor and image.dim() == 4:
# If it's a batch with single image, remove batch dimension
if image.shape[0] == 1:
image = image.squeeze(0)
else:
raise ValueError("defend_single expects a single image, got batch")
# Create multiscale pyramid
pyramid = {}
for scale in self.scales:
pyramid[scale] = self.transform(image, scale)
# Apply DDA blocks
for _ in range(self.num_dda_blocks):
pyramid = self.dda_block(pyramid)
# Convert back to original scale and average
original_scale_images = []
for scale, scaled_image in pyramid.items():
if scale != 1.0:
restored = self.inverse_transform(scaled_image, scale)
# Resize to match original if needed
if is_tensor:
# Ensure proper dimensions for interpolation
if restored.dim() == 3:
restored_4d = restored.unsqueeze(0)
else:
restored_4d = restored
if restored.shape[-2:] != image.shape[-2:]:
restored_4d = F.interpolate(
restored_4d,
size=(image.shape[-2], image.shape[-1]),
mode='bilinear',
align_corners=False
)
if restored.dim() == 3:
restored = restored_4d.squeeze(0)
else:
restored = restored_4d
restored = restored.to(device)
else:
if restored.shape != image.shape:
restored = cv2.resize(
restored,
(image.shape[1], image.shape[0]),
interpolation=cv2.INTER_LINEAR
)
original_scale_images.append(restored)
else:
original_scale_images.append(scaled_image)
# Average the images
if is_tensor:
defended = torch.zeros_like(image, device=device) # Fix: specify device
for img in original_scale_images:
defended = defended + img.to(device) # Ensure all on same device
defended = defended / len(original_scale_images)
else:
defended = np.zeros_like(image)
for img in original_scale_images:
defended = defended + img
defended = defended / len(original_scale_images)
return defended
References
- Wang Y. et al. (2023). Reversing Skin Cancer Adversarial Examples by Multiscale Diffusive and Denoising Aggregation Mechanism. Computers in Biology and Medicine.
- Finlayson S.G. et al. (2019). Adversarial Attacks on Medical Machine Learning. Science.
- ISIC 2019 Dataset: Challenge Archive