Medical image segmentation is the cornerstone of modern diagnostics and treatment planning. From pinpointing tumor boundaries to mapping cardiac structures, its precision directly impacts patient outcomes. Yet, a critical bottleneck persists: the massive annotation burden. Manual labeling demands hours of expert time per scan, creating a severe shortage of labeled data that throttles AI’s potential. Enter semi-supervised learning (SSL) – the beacon of hope promising to leverage abundant unlabeled scans. While frameworks like FixMatch showed initial promise, their limitations in capturing complex medical semantics hindered true progress. SemSim emerges as the solution, redefining SSL for medical images through the lens of semantic similarity. This breakthrough not only slashes annotation needs but achieves unprecedented accuracy, setting a new standard for the field.
The Annotation Crisis & FixMatch’s Shortcomings in Medicine
Medical imaging generates petabytes of data, yet only a tiny fraction is expertly labeled. This scarcity cripples fully supervised deep learning models. SSL techniques like FixMatch offered a path forward by leveraging weak-to-strong consistency regularization:
- The FixMatch Process (Simplified):
- Apply a weak augmentation (e.g., slight rotation) to an unlabeled image, generate a prediction (
pw). - Apply a strong augmentation (e.g., color distortion, CutMix) to the same image, generate another prediction (
ps). - Use high-confidence pixels from
pw as pseudo-labels to superviseps.
- Apply a weak augmentation (e.g., slight rotation) to an unlabeled image, generate a prediction (
While successful in simpler classification tasks, FixMatch stumbles on the intricate demands of medical segmentation:
- Intra-Image Semantic Fragmentation (The “Island Problem”):
- FixMatch enforces pixel-wise consistency but ignores crucial contextual dependencies within the image.
- Features representing the same anatomical structure (e.g., part of the left ventricle) might exhibit high similarity yet receive inconsistent predictions due to local noise or weak boundaries.
- This leads to disconnected segments, holes, and incomplete structures – clinically unacceptable results. (Visualized as discontinuous features in Fig. 3 left column).
- Cross-Image Semantic Discrepancy (The “Distribution Drift”):
- FixMatch treats each unlabeled image in isolation.
- It fails to leverage the inherent anatomical similarity across different patients’ scans (e.g., all healthy livers share common features).
- With limited labeled data, the model learns a skewed class distribution. Predictions on unlabeled data then exhibit a significant mismatch compared to the true underlying distribution learned from labeled data. (Visualized as diverging kernel density plots in Fig. 3 right column).
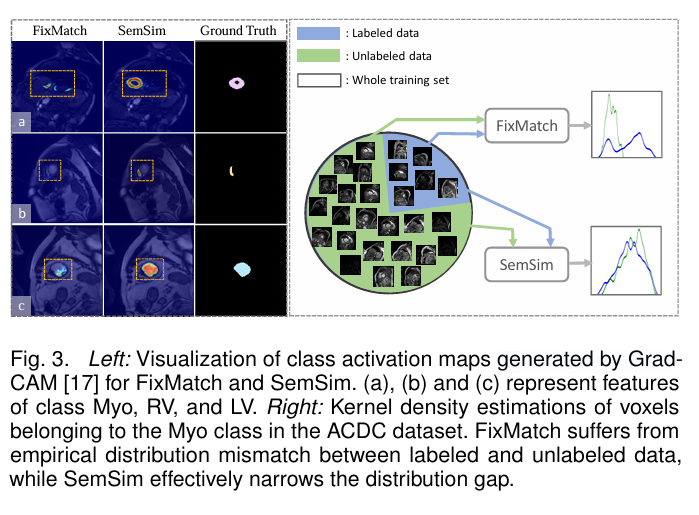
These limitations meant that simply applying FixMatch to medical images was insufficient. A fundamentally different approach, grounded in medical image semantics, was needed. SemSim provides this paradigm shift.
SemSim: Semantic Similarity as the Guiding Principle
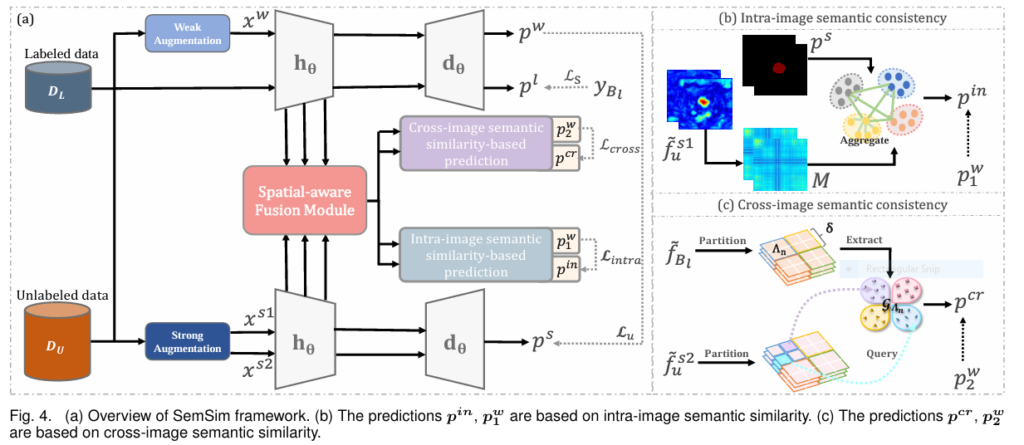
SemSim revolutionizes SSL for medical segmentation by embedding semantic similarity at its core. It retains FixMatch’s efficient weak-to-strong consistency backbone but introduces two transformative consistency constraints and a powerful feature fusion engine.
Core Innovations:
- Intra-Image Semantic Consistency: Context is King
- Problem Addressed: Fragmented predictions, lack of spatial coherence.
- Solution: Leverage feature-level affinity maps to explicitly model relationships between pixels/regions within the same image.
- How it Works:
- Extract robust multi-scale features using the Spatial-aware Fusion Module (SFM – see below).
- Compute an affinity matrix (
M) where each elementM(k1, k2)measures the cosine similarity between features at positionsk1andk2(Eq. 4). - Refine the initial strong augmentation prediction (
ps) by integrating this affinity information:pin = I(ps) + I(ps) . M(Eq. 5). This essentially “spreads” high-affinity (similar) regions towards the same label. - Apply a consistency loss (
Lintra) between the refined prediction for the strong view (pin) and a similarly refined prediction for the weak view (pw1).
- Impact: Forces the model to understand that pixels with similar semantic features should have the same label, leading to smoother, more continuous, and complete segmentations. (Fig. 3 left shows improved feature continuity).
- Cross-Image Semantic Consistency: Learning from the Collective
- Problem Addressed: Distribution mismatch between limited labeled data and unlabeled data.
- Solution: Bridge labeled and unlabeled data by dynamically querying reliable class prototypes derived from labeled data.
- How it Works:
- Local Prototype Extraction: Avoid averaging noisy background features! For labeled images, partition the feature map into sub-regions (e.g., 2×2 grids). Within each sub-region
Λn, compute a local class prototype (GcΛn) by averaging features belonging to classc(Eq. 7). This captures spatially consistent anatomical patterns. - Dynamic Feature Querying: For an unlabeled image’s feature (
fus2), partition it similarly. For each sub-region in the unlabeled image, compute its cosine similarity (mΛn) to the corresponding sub-region’s prototypes from the entire labeled batch (Eq. 8). - Prediction Generation: Convert these similarity scores across all sub-regions and labeled images into a robust prediction (
p_cr) for the unlabeled image’s strong view (Eq. 9). - Uncertainty-Aware Constraint: Calculate the uncertainty (
U) ofpcr based on prediction variance across the labeled batch (Eqs. 10-12). Apply a consistency loss (Lcross) betweenpcr and the corresponding weak-view prediction (pw2), weighted by this uncertainty (Eq. 13). Low confidence predictions contribute less to learning.
- Local Prototype Extraction: Avoid averaging noisy background features! For labeled images, partition the feature map into sub-regions (e.g., 2×2 grids). Within each sub-region
- Impact: Actively transfers reliable knowledge from labeled data to unlabeled data, aligning class distributions and enabling the model to learn consistent anatomical representations even with scarce labels. (Fig. 3 right shows narrowed distribution gap).
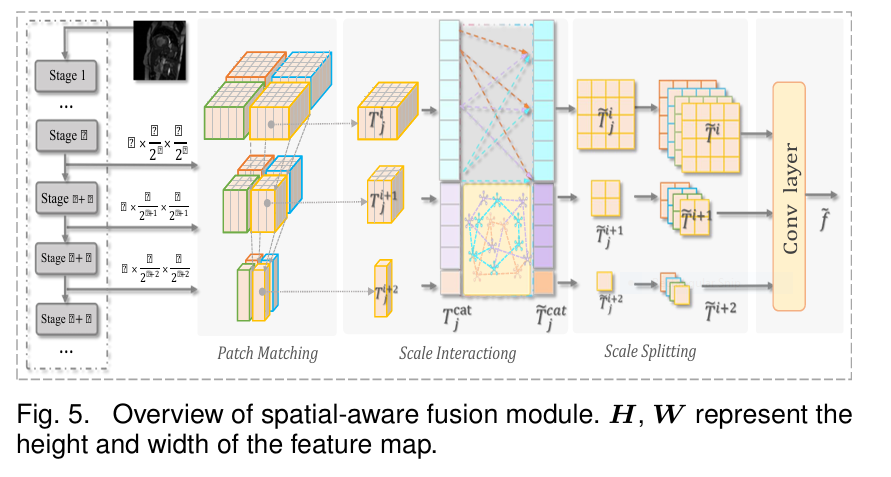
- Spatial-Aware Fusion Module (SFM): Powering Reliable Similarity
- The Need: Accurate affinity maps (
M) and feature querying depend on robust, multi-scale feature representations. Medical structures exist at various scales. - Solution: A lightweight Transformer-based module for efficient cross-scale fusion, respecting spatial coherence.
- How it Works :
- Patch Matching: Instead of naively concatenating all scales (computationally expensive!), exploit inherent spatial hierarchies. For a patch at scale
i, find its corresponding (down-sampled) patches at scalesi+1andi+2that cover the same image region (e.g., yellow bounding boxes). - Scale Interacting: Concatenate features from these corresponding patches (Eq. 14). Apply efficient Multi-head Self-Attention (MSA) and MLP layers only within these matched groups (Eq. 15).
- Scale Splitting & Fusion: Split the enhanced features back to their respective scales, interpolate to a uniform size, and fuse via convolution (Eqs. 16, 17).
- Patch Matching: Instead of naively concatenating all scales (computationally expensive!), exploit inherent spatial hierarchies. For a patch at scale
- Impact: Generates powerful, context-rich features by fusing local details and global semantics efficiently. Crucially enables reliable semantic similarity calculation for both intra- and cross-image constraints. Boosts performance with minimal parameter overhead (+0.79M).
- The Need: Accurate affinity maps (
The Unified SemSim Framework: SemSim processes an unlabeled image through dual strong perturbation streams (xa1, xa2). Alongside the standard FixMatch loss (Lu), it calculates:
Lintra usingxa1and the weak view.Lcross usingxa2and the weak view.
The total unsupervised loss is a weighted sum (Eq. 18), combined with the supervised loss on labeled data (Eq. 19).
Proven Superiority: SemSim Outperforms the State-of-the-Art for Medical Image Segmentation
Rigorous evaluation across three major public medical segmentation benchmarks (ACDC – Cardiac MRI, ISIC – Skin Lesions, PROMISE12 – Prostate MRI) under extreme label scarcity (1%, 3%, 5%, 7%, 10% labeled) demonstrates SemSim’s dominance:
- ACDC (Cardiac Structures – Table I):
- 1% Labels (1 patient): SemSim (DSC: 87.2%, 95HD: 1.8mm) crushed competitors. FixMatch: 72.2%/22.8mm, CPC-SAM: 85.6%/9.2mm. +1.6% DSC, -7.4mm 95HD over CPC-SAM.
- 5% Labels (3 patients): SemSim (88.8%/1.9mm) beat CPC-SAM (88.0%/5.8mm).
- 10% Labels (7 patients): SemSim (89.6%/2.3mm) outperformed all, including BCP-Net (88.9%/4.0mm) and CPC-SAM (89.0%/3.1mm).
- Efficiency: SemSim achieved SOTA with only 2.6 million parameters vs. CPC-SAM’s 93.75M.
- ISIC (Skin Lesions – Table II):
- 3% Labels (55 images): SemSim (DSC: 77.4%, 95HD: 18.9mm) beat BCP-Net (76.9%/24.5mm) and FixMatch (75.6%/36.7mm).
- 10% Labels (181 images): SemSim (80.2%/13.3mm) again led, surpassing BCP-Net (79.4%/18.7mm) and FixMatch (78.3%/15.5mm).
- PROMISE12 (Prostate – Table III):
- 3% Labels: SemSim (75.8%/1.6mm) significantly outperformed MedFCT (72.5%/2.2mm) and BCP-Net (70.6%/8.8mm).
- 7% Labels: SemSim (78.4%/1.3mm) maintained leadership over MedFCT (76.0%/1.8mm) and BCP-Net (77.2%/1.4mm).
Visual Evidence (Fig. 6): Qualitative results on ACDC and ISIC with only 10% labeled data show SemSim produces segmentations remarkably close to the ground truth, with sharper boundaries, fewer holes, and better structural integrity compared to FixMatch, MedFCT, CnT-B, BCP-Net, CPC-SAM, and DiffRect.
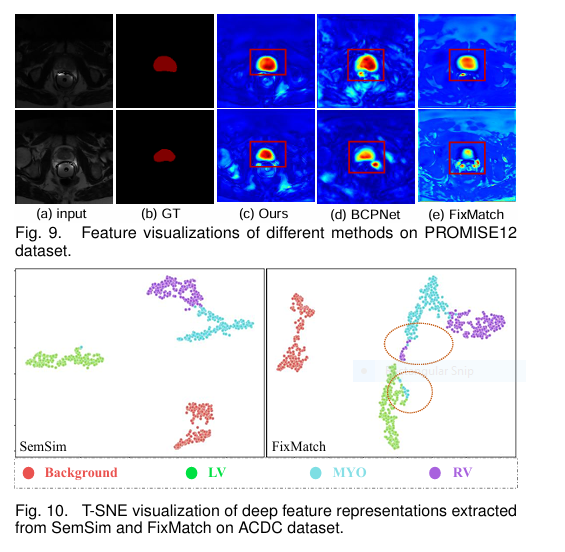
Why SemSim Wins (Figs. 9 & 10):
- Intra-Image Cohesion: Class activation maps (Fig. 9) reveal SemSim produces more complete and connected activation regions compared to FixMatch and BCP-Net.
- Cross-Image Alignment: T-SNE visualizations (Fig. 10) show features learned by SemSim form tighter, more distinct clusters per class compared to FixMatch, proving its success in learning consistent and separable class distributions.
Beyond Benchmarks: The Real-World Impact of SemSim
SemSim isn’t just a research triumph; it’s a practical solution for healthcare:
- Dramatically Reduced Annotation Costs: Achieve high accuracy with 90-99% fewer labeled scans. This makes deploying AI segmentation feasible for hospitals and labs without massive annotation budgets.
- Enhanced Diagnostic Precision: More accurate and continuous segmentations translate directly to better measurements, early disease detection, and precise treatment planning (e.g., tumor volume assessment, cardiac function analysis).
- Accelerated Research: Researchers can prototype and validate new segmentation models faster by leveraging existing unlabeled datasets.
- Lightweight & Efficient: With its modest parameter count, SemSim is suitable for deployment in various clinical computing environments.
If you’re Interested in semi-supervised learning with advance methods, you may also find this article helpful: Uncertainty Beats Confidence in semi-supervised learning
The Future of Medical Imaging is Semi-Supervised
SemSim represents a fundamental leap forward in semi-supervised medical image segmentation. By moving beyond naive pixel consistency and deeply integrating semantic similarity – both within single images and across the dataset – it unlocks unprecedented performance with minimal labeled data. Its innovative components, the intra-image affinity refinement, cross-image prototype querying, and spatial-aware feature fusion, provide a robust blueprint for future SSL research in medicine.
Ready to harness the power of semantic similarity for your medical imaging challenges?
- Researchers: Dive into the original SemSim paper for implementation details and extend this groundbreaking work. Explore integrating SemSim principles with foundation models like SAM.
- Clinicians & Labs: Partner with AI developers to implement SemSim-based solutions, drastically reducing your annotation burden while improving segmentation quality for critical diagnostic tasks.
- Developers: Integrate the SemSim framework into your medical imaging pipelines to deliver state-of-the-art segmentation accuracy efficiently.
Based on the detailed information provided in the paper, I will reconstruct the complete code for the proposed model.
import torch
import torch.nn as nn
import torch.nn.functional as F
from einops import rearrange, repeat
class SpatialAwareFusion(nn.Module):
"""Spatial-Aware Fusion Module (SFM)"""
def __init__(self, in_channels, num_scales=3, patch_size=16):
super().__init__()
self.patch_size = patch_size
self.num_scales = num_scales
self.norm = nn.LayerNorm(in_channels)
self.attn = nn.MultiheadAttention(in_channels, num_heads=8)
self.mlp = nn.Sequential(
nn.Linear(in_channels, 4 * in_channels),
nn.GELU(),
nn.Linear(4 * in_channels, in_channels)
)
self.fusion_conv = nn.Sequential(
nn.Conv2d(in_channels * num_scales, in_channels, 3, padding=1),
nn.BatchNorm2d(in_channels),
nn.ReLU(inplace=True)
)
def forward(self, features):
# features: list of [B, C, H, W] at different scales
B, C, H, W = features[0].shape
device = features[0].device
# Create patch grid
patches = []
for feat in features:
p = F.unfold(feat, kernel_size=self.patch_size, stride=self.patch_size)
patches.append(p) # [B, C*P, N]
# Process corresponding patches across scales
enhanced_patches = []
for i in range(patches[0].shape[-1]):
patch_group = []
for p in patches:
patch = p[..., i].view(B, C, -1).permute(2, 0, 1) # [P, B, C]
patch_group.append(patch)
# Concatenate corresponding patches
cat_patch = torch.cat(patch_group, dim=0) # [num_scales*P, B, C]
cat_patch = self.norm(cat_patch)
# Self-attention
attn_out, _ = self.attn(cat_patch, cat_patch, cat_patch)
attn_out = attn_out + cat_patch
mlp_out = self.mlp(self.norm(attn_out))
enhanced_patches.append(mlp_out + attn_out)
# Reconstruct feature maps
enhanced_features = []
for scale_idx in range(self.num_scales):
scale_patches = []
for i in range(len(enhanced_patches)):
start_idx = scale_idx * (self.patch_size ** 2)
end_idx = (scale_idx + 1) * (self.patch_size ** 2)
patch = enhanced_patches[i][start_idx:end_idx] # [P, B, C]
patch = patch.permute(1, 2, 0).view(B, C, self.patch_size, self.patch_size)
scale_patches.append(patch)
# Fold patches back to feature map
scale_feature = F.fold(
torch.cat(scale_patches, dim=1),
output_size=(H, W),
kernel_size=self.patch_size,
stride=self.patch_size
)
enhanced_features.append(scale_feature)
# Fuse multi-scale features
fused = self.fusion_conv(torch.cat(enhanced_features, dim=1))
return fused
class SemSim(nn.Module):
"""SemSim: Semi-supervised Medical Image Segmentation Framework"""
def __init__(self, num_classes, backbone_channels=[64, 128, 256, 512]):
super().__init__()
self.num_classes = num_classes
# Backbone (UNet-like encoder-decoder)
self.encoder = nn.ModuleList([
nn.Sequential(
nn.Conv2d(3, backbone_channels[0], 3, padding=1),
nn.BatchNorm2d(backbone_channels[0]),
nn.ReLU(inplace=True),
nn.Conv2d(backbone_channels[0], backbone_channels[0], 3, padding=1),
nn.BatchNorm2d(backbone_channels[0]),
nn.ReLU(inplace=True)
),
nn.Sequential(
nn.MaxPool2d(2),
nn.Conv2d(backbone_channels[0], backbone_channels[1], 3, padding=1),
nn.BatchNorm2d(backbone_channels[1]),
nn.ReLU(inplace=True),
nn.Conv2d(backbone_channels[1], backbone_channels[1], 3, padding=1),
nn.BatchNorm2d(backbone_channels[1]),
nn.ReLU(inplace=True)
),
nn.Sequential(
nn.MaxPool2d(2),
nn.Conv2d(backbone_channels[1], backbone_channels[2], 3, padding=1),
nn.BatchNorm2d(backbone_channels[2]),
nn.ReLU(inplace=True),
nn.Conv2d(backbone_channels[2], backbone_channels[2], 3, padding=1),
nn.BatchNorm2d(backbone_channels[2]),
nn.ReLU(inplace=True)
),
nn.Sequential(
nn.MaxPool2d(2),
nn.Conv2d(backbone_channels[2], backbone_channels[3], 3, padding=1),
nn.BatchNorm2d(backbone_channels[3]),
nn.ReLU(inplace=True),
nn.Conv2d(backbone_channels[3], backbone_channels[3], 3, padding=1),
nn.BatchNorm2d(backbone_channels[3]),
nn.ReLU(inplace=True)
)
])
# Spatial-Aware Fusion Modules
self.sfm2 = SpatialAwareFusion(backbone_channels[1])
self.sfm3 = SpatialAwareFusion(backbone_channels[2])
self.sfm4 = SpatialAwareFusion(backbone_channels[3])
# Decoder
self.decoder = nn.ModuleList([
nn.Sequential(
nn.ConvTranspose2d(backbone_channels[3], backbone_channels[2], 2, stride=2),
nn.BatchNorm2d(backbone_channels[2]),
nn.ReLU(inplace=True)
),
nn.Sequential(
nn.ConvTranspose2d(backbone_channels[2], backbone_channels[1], 2, stride=2),
nn.BatchNorm2d(backbone_channels[1]),
nn.ReLU(inplace=True)
),
nn.Sequential(
nn.ConvTranspose2d(backbone_channels[1], backbone_channels[0], 2, stride=2),
nn.BatchNorm2d(backbone_channels[0]),
nn.ReLU(inplace=True)
)
])
# Prediction heads
self.seg_head = nn.Conv2d(backbone_channels[0], num_classes, 1)
# Feature projection for affinity computation
self.feature_proj = nn.Sequential(
nn.Conv2d(backbone_channels[0], 128, 1),
nn.BatchNorm2d(128),
nn.ReLU(inplace=True)
)
def forward_encoder(self, x):
features = []
for i, layer in enumerate(self.encoder):
x = layer(x)
if i > 0: # Skip first layer for SFM
features.append(x)
# Apply SFM at stages 2, 3, 4
f2 = self.sfm2([features[0]])
f3 = self.sfm3([features[1]])
f4 = self.sfm4([features[2]])
return [f2, f3, f4], x # Multi-scale features and final feature
def forward_decoder(self, features):
x = features[-1]
for i, dec_layer in enumerate(self.decoder):
x = dec_layer(x)
if i < len(features) - 1:
x = torch.cat([x, features[-(i+2)]], dim=1)
return x
def forward(self, x):
# Encoder
features, final_feat = self.forward_encoder(x)
# Decoder
dec_out = self.forward_decoder(features)
# Segmentation prediction
seg_pred = self.seg_head(dec_out)
# Enhanced features for affinity
proj_feat = self.feature_proj(dec_out)
return seg_pred, proj_feat
class SemSimFramework:
"""Complete SemSim Training Framework"""
def __init__(self, num_classes, device='cuda'):
self.model = SemSim(num_classes).to(device)
self.device = device
self.num_classes = num_classes
# Loss weights
self.lambda_s = 1.0
self.lambda_u = 1.0
self.lambda_intra = 0.25
self.lambda_cross = 0.25
# Confidence threshold
self.conf_threshold = 0.95
# Optimizer
self.optimizer = torch.optim.SGD(
self.model.parameters(),
lr=0.01,
momentum=0.9,
weight_decay=1e-4
)
def compute_affinity(self, features):
"""Compute intra-image affinity map"""
# features: [B, C, H, W]
B, C, H, W = features.shape
features_flat = rearrange(features, 'b c h w -> b (h w) c')
# Normalize
features_norm = F.normalize(features_flat, p=2, dim=-1)
# Compute affinity
affinity = torch.bmm(features_norm, features_norm.transpose(1, 2))
affinity = F.softmax(affinity, dim=-1)
return affinity # [B, HW, HW]
def intra_image_consistency(self, weak_pred, weak_feat, strong_pred, strong_feat):
"""Intra-image semantic consistency"""
# Compute affinity maps
weak_affinity = self.compute_affinity(weak_feat)
strong_affinity = self.compute_affinity(strong_feat)
# Reshape predictions
B, C, H, W = weak_pred.shape
weak_pred_flat = rearrange(weak_pred, 'b c h w -> b c (h w)')
strong_pred_flat = rearrange(strong_pred, 'b c h w -> b c (h w)')
# Refine predictions using affinity
refined_weak = weak_pred_flat + torch.bmm(weak_pred_flat, weak_affinity)
refined_strong = strong_pred_flat + torch.bmm(strong_pred_flat, strong_affinity)
# Reshape back
refined_weak = rearrange(refined_weak, 'b c (h w) -> b c h w', h=H, w=W)
refined_strong = rearrange(refined_strong, 'b c (h w) -> b c h w', h=H, w=W)
# Confidence masking
conf_mask = (torch.amax(weak_pred, dim=1) > self.conf_threshold).float()
# Loss calculation
loss = dice_loss(refined_strong, refined_weak.detach(), mask=conf_mask)
return loss
def cross_image_consistency(self, labeled_batch, unlabeled_strong_pred, unlabeled_strong_feat):
"""Cross-image semantic consistency"""
lbl_imgs, lbl_masks = labeled_batch
B_u, C, H, W = unlabeled_strong_pred.shape
B_l = lbl_imgs.shape[0]
# Extract prototypes from labeled batch
with torch.no_grad():
_, lbl_feat = self.model(lbl_imgs)
lbl_feat = F.normalize(lbl_feat, p=2, dim=1)
# Split feature maps into sub-regions (2x2 grid)
grid_size = 2
unl_feat_subs = []
lbl_feat_subs = []
lbl_mask_subs = []
for i in range(grid_size):
for j in range(grid_size):
h_start = i * H // grid_size
h_end = (i+1) * H // grid_size
w_start = j * W // grid_size
w_end = (j+1) * W // grid_size
unl_feat_subs.append(unlabeled_strong_feat[:, :, h_start:h_end, w_start:w_end])
lbl_feat_subs.append(lbl_feat[:, :, h_start:h_end, w_start:w_end])
lbl_mask_subs.append(lbl_masks[:, :, h_start:h_end, w_start:w_end])
# Compute similarity and predictions
pred_parts = []
uncertainties = []
for idx, (unl_sub, lbl_sub, lbl_mask_sub) in enumerate(zip(
unl_feat_subs, lbl_feat_subs, lbl_mask_subs
)):
# Flatten spatial dimensions
unl_flat = rearrange(unl_sub, 'b c h w -> b (h w) c')
lbl_flat = rearrange(lbl_sub, 'b c h w -> b (h w) c')
lbl_mask_flat = rearrange(lbl_mask_sub, 'b c h w -> b (h w) c')
# Compute class prototypes per labeled image
prototypes = []
for cls in range(self.num_classes):
cls_mask = (lbl_mask_flat == cls).float()
if cls_mask.sum() > 0:
cls_proto = (lbl_flat * cls_mask).sum(1) / cls_mask.sum(1)
else:
cls_proto = torch.zeros(B_l, C, device=self.device)
prototypes.append(cls_proto)
prototypes = torch.stack(prototypes, dim=1) # [B_l, num_classes, C]
# Compute similarity
similarity = torch.einsum('bqc,bkc->bqk', unl_flat, prototypes) # [B_u, Q, B_l, num_classes]
similarity = rearrange(similarity, 'b q k c -> b k c q')
# Compute prediction
pred_part = F.softmax(similarity.mean(dim=1), dim=1) # [B_u, num_classes, Q]
pred_parts.append(rearrange(pred_part, 'b c (h w) -> b c h w',
h=unl_sub.shape[2], w=unl_sub.shape[3]))
# Compute uncertainty
avg_similarity = similarity.mean(dim=1)
entropy = -torch.sum(avg_similarity * torch.log(avg_similarity + 1e-10), dim=1)
uncertainties.append(entropy)
# Combine sub-region predictions
pred_full = torch.zeros(B_u, self.num_classes, H, W, device=self.device)
uncertainty_full = torch.zeros(B_u, H, W, device=self.device)
sub_idx = 0
for i in range(grid_size):
for j in range(grid_size):
h_start = i * H // grid_size
h_end = (i+1) * H // grid_size
w_start = j * W // grid_size
w_end = (j+1) * W // grid_size
pred_full[:, :, h_start:h_end, w_start:w_end] = pred_parts[sub_idx]
uncertainty_full[:, h_start:h_end, w_start:w_end] = uncertainties[sub_idx]
sub_idx += 1
# Uncertainty weighting
weights = torch.exp(-uncertainty_full)
# Loss calculation
conf_mask = (torch.amax(unlabeled_strong_pred, dim=1) > self.conf_threshold).float()
loss = weighted_dice_loss(
pred_full,
unlabeled_strong_pred.detach(),
mask=conf_mask,
weights=weights
)
return loss
def train_step(self, labeled_batch, unlabeled_batch):
"""Perform a single training step"""
lbl_imgs, lbl_masks = labeled_batch
unl_imgs_weak, unl_imgs_strong1, unl_imgs_strong2 = unlabeled_batch
# Move to device
lbl_imgs, lbl_masks = lbl_imgs.to(self.device), lbl_masks.to(self.device)
unl_imgs_weak = unl_imgs_weak.to(self.device)
unl_imgs_strong1 = unl_imgs_strong1.to(self.device)
unl_imgs_strong2 = unl_imgs_strong2.to(self.device)
# Forward pass for labeled data
lbl_pred, _ = self.model(lbl_imgs)
sup_loss = 0.5 * (F.cross_entropy(lbl_pred, lbl_masks) + dice_loss(lbl_pred, lbl_masks))
# Forward pass for unlabeled data - weak augmentation
with torch.no_grad():
weak_pred, weak_feat = self.model(unl_imgs_weak)
# Forward pass for unlabeled data - strong augmentations
strong_pred1, strong_feat1 = self.model(unl_imgs_strong1)
strong_pred2, strong_feat2 = self.model(unl_imgs_strong2)
# FixMatch consistency loss
conf_mask = (torch.amax(weak_pred, dim=1) > self.conf_threshold).float()
fixmatch_loss = dice_loss(strong_pred1, weak_pred.detach(), mask=conf_mask)
# Intra-image consistency loss
intra_loss = self.intra_image_consistency(
weak_pred, weak_feat,
strong_pred1, strong_feat1
)
# Cross-image consistency loss
cross_loss = self.cross_image_consistency(
(lbl_imgs, lbl_masks),
strong_pred2,
strong_feat2
)
# Total loss
total_loss = (
self.lambda_s * sup_loss +
self.lambda_u * fixmatch_loss +
self.lambda_intra * intra_loss +
self.lambda_cross * cross_loss
)
# Backward pass
self.optimizer.zero_grad()
total_loss.backward()
self.optimizer.step()
return {
'total_loss': total_loss.item(),
'sup_loss': sup_loss.item(),
'fixmatch_loss': fixmatch_loss.item(),
'intra_loss': intra_loss.item(),
'cross_loss': cross_loss.item()
}
# Helper functions
def dice_loss(pred, target, mask=None, smooth=1e-5):
"""Dice loss for segmentation"""
pred = F.softmax(pred, dim=1)
target = F.one_hot(target, num_classes=pred.shape[1]).permute(0, 3, 1, 2).float()
intersection = torch.sum(pred * target, dim=(2, 3))
union = torch.sum(pred, dim=(2, 3)) + torch.sum(target, dim=(2, 3))
dice = (2. * intersection + smooth) / (union + smooth)
loss = 1 - dice
if mask is not None:
loss = loss * mask.mean(dim=(1,2))
return loss.mean()
def weighted_dice_loss(pred, target, mask=None, weights=None, smooth=1e-5):
"""Weighted Dice loss"""
pred = F.softmax(pred, dim=1)
target = F.softmax(target, dim=1)
intersection = torch.sum(pred * target, dim=(2, 3))
union = torch.sum(pred, dim=(2, 3)) + torch.sum(target, dim=(2, 3))
dice = (2. * intersection + smooth) / (union + smooth)
loss = 1 - dice
if mask is not None:
loss = loss * mask.mean(dim=(1,2))
if weights is not None:
weights = weights.mean(dim=(1,2))
loss = loss * weights
return loss.mean()# Initialize framework
model = SemSimFramework(num_classes=3, device='cuda')
# Training loop
for epoch in range(300):
for labeled_batch, unlabeled_batch in train_loader:
# labeled_batch: (images, masks)
# unlabeled_batch: (weak_aug, strong_aug1, strong_aug2)
losses = model.train_step(labeled_batch, unlabeled_batch)
# Log losses, update LR, etc.
# Inference
def predict(image):
model.model.eval()
with torch.no_grad():
pred, _ = model.model(image)
return torch.softmax(pred, dim=1)
Pingback: CMKD: Slash 99% Storage Costs & Dominate UDA Challenges - aitrendblend.com