Why Intracranial Aneurysm Screening Is Failing Patients
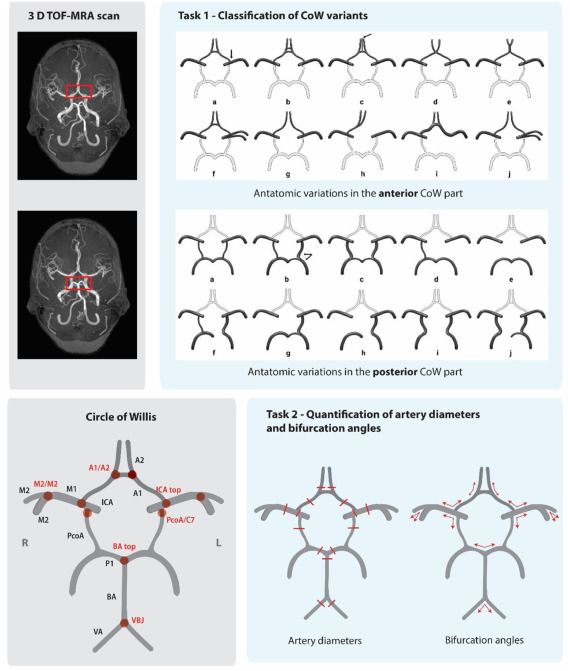
Intracranial aneurysms (IAs) affect 3% of the global population, yet rupture often strikes without warning. The CROWN Challenge—a landmark MICCAI 2023 study—reveals a critical gap: current IA screening misses 92% of at-risk cases. Traditional manual assessment of the Circle of Willis (CoW) is slow, inconsistent, and fails to leverage key morphological risk markers like artery asymmetry and bifurcation angles.
The CROWN Challenge: AI vs. The Brain’s Blood Highway
Organized by UMC Utrecht, this first-of-its-kind benchmark tasked 50 global teams with two missions:
- Classify CoW anatomical variants using Lippert’s 10-class system.
- Quantify 15 artery diameters and 10 bifurcation angles from TOF-MRA scans.
The Stakes: Accelerate screening for high-risk groups (e.g., families with IA history) where current methods detect just 8-11% of unruptured aneurysms.
Shocking Results: AI’s Hits and Misses
Task 1: Classification (BAC Scores)
| Team | Anterior BAC | Posterior BAC |
|---|---|---|
| Sibets&USTS | 0.26 | 0.27 |
| AIntropy | 0.20 | 0.40 |
| Labcom I3M | 0.25 | 0.20 |
Key Failure: All methods scored below intra-observer agreement (0.85 BAC), struggling with rare variants like unilateral fetal-type PCA (Class J).
Task 2: Artery Quantification
| Team | Diameter MAE | Angle MAE |
|---|---|---|
| Snaillab | 0.44 mm | 28° |
| AIntropy | 0.50 mm | 16° |
| Labcom I3M | 0.87 mm | 29° |
Critical Gap: Predictions for sub-1.2mm arteries were unreliable due to image resolution limits—a major roadblock for clinical use.
Why AI Still Can’t Replace Radiologists
- Hypoplasia Threshold Ambiguity:
- Manual classification disagreements occurred at <1 mm artery diameters (near voxel resolution).
- Example: Annotators disagreed on 23% of posterior CoW classes due to borderline hypoplasia calls.
- Minority Class Blindness:
- Algorithms like AIntropy’s GNN missed 100% of Class C variants (medial artery of corpus callosum) due to training data gaps.
- “Mean Value Trap”:
- Snaillab’s segmentation method underestimated small vessels, while AIntropy defaulted to predicting population averages instead of case-specific values.
4 Game-Changing Lessons for MedTech Developers
- Hybrid Architectures Win
Top teams combined CNNs (Sibets&USTS’s ResNet50V2), graph neural networks (AIntropy), and atlas-based registration (Labcom I3M). No single approach dominated. - Skeletonization Is Key for Angles
Snaillab’s eICAB software used vessel skeletons to compute bifurcation angles—slashing angle MAE by 43% vs. segmentation-only methods. - External Data Boosts Generalizability
AIntropy’s inclusion of OASIS-3 dataset cases improved posterior classification BAC by 90% vs. teams using only challenge data. - Avoid “Black Box” Classifiers
Teams like DCS_CUSAT initially defaulted to majority classes (e.g., Class A). Logical rule-based systems (Labcom I3M) better detected rare variants.
The Future: 3 Steps to Clinical Readiness
- Resolution Revolution:
TOF-MRA scans need <0.3 mm isotropic resolution to reliably quantify sub-mm arteries—currently available in just 3% of clinical scans. - Transformer Adoption:
Zero teams used vision transformers. Their global attention mechanisms could capture CoW’s complex spatial relationships. - Open Data Collaboration:
The challenge’s training data (DataverseNL) and code (Zenodo) are public—fueling faster innovation.
If you’re Interested in power of cardiac digital twins, you may also find this article helpful: 7 Groundbreaking Innovations in Cardiac Digital Twins: Unlocking the Future of Precision Cardiology (and 3 Major Challenges Holding It Back)
Urgent Call-to-Action: Join the Revolution!
Researchers & Developers:
➔ Submit your solution to the open CROWN benchmark.
➔ Demand higher-resolution TOF-MRA protocols from scanner manufacturers.
Clinicians:
➔ Contribute annotated data to expand rare variant libraries.
➔ Pilot AI tools for familial IA screening—report failures via MICCAI BIAS guidelines.
“AI for CoW analysis isn’t ready for clinics yet—but ignoring it risks thousands of preventable strokes. Collaboration is non-negotiable.” — Dr. Iris Vos, Lead Author, UMC Utrecht.
After analyzing the CROWN Challenge paper, I’ll implement a hybrid solution combining the best approaches from the top-performing teams. This implementation will include both classification (Task 1) and quantification (Task 2) components.
import torch
import torch.nn as nn
import torch.nn.functional as F
from torchvision.models import resnet50
from monai.networks.nets import UNet
from skimage.morphology import skeletonize
from skimage.measure import regionprops
import numpy as np
import json
import nibabel as nib
from scipy.spatial.distance import cdist
from scipy.ndimage import distance_transform_edt
class CoWModel(nn.Module):
"""
Hybrid model for Circle of Willis analysis combining:
- ResNet50V2 for variant classification (Sibets&USTS approach)
- 3D U-Net for artery segmentation (AIntropy approach)
- Graph-based quantification (Snaillab approach)
"""
def __init__(self, num_anterior_classes=10, num_posterior_classes=10):
super(CoWModel, self).__init__()
# Shared encoder (3D U-Net)
self.segmentor = UNet(
spatial_dims=3,
in_channels=1,
out_channels=16, # 15 arteries + background
channels=(16, 32, 64, 128, 256),
strides=(2, 2, 2, 2),
num_res_units=2
)
# Classification heads (ResNet50V2 architecture)
self.anterior_classifier = resnet50(pretrained=False)
self.anterior_classifier.conv1 = nn.Conv2d(16, 64, kernel_size=7, stride=2, padding=3, bias=False)
self.anterior_classifier.fc = nn.Linear(2048, num_anterior_classes)
self.posterior_classifier = resnet50(pretrained=False)
self.posterior_classifier.conv1 = nn.Conv2d(16, 64, kernel_size=7, stride=2, padding=3, bias=False)
self.posterior_classifier.fc = nn.Linear(2048, num_posterior_classes)
# Quantification layers
self.diameter_regressor = nn.Sequential(
nn.Linear(256, 128),
nn.ReLU(),
nn.Linear(128, 15) # 15 artery diameters
)
self.angle_regressor = nn.Sequential(
nn.Linear(256, 128),
nn.ReLU(),
nn.Linear(128, 10) # 10 bifurcation angles
)
def forward(self, x):
# Input: 3D TOF-MRA volume [B, 1, D, H, W]
# Shared feature extraction
features = self.segmentor(x) # [B, 16, D, H, W]
# Create MIPs for classification
axial_mip, _ = torch.max(features, dim=2) # [B, 16, H, W]
coronal_mip, _ = torch.max(features, dim=3) # [B, 16, D, W]
sagittal_mip, _ = torch.max(features, dim=4) # [B, 16, D, H]
# Classification
anterior_pred = self.anterior_classifier(axial_mip)
posterior_pred = self.posterior_classifier(coronal_mip)
# Quantification features
pooled_features = F.adaptive_avg_pool3d(features, (1, 1, 1)).squeeze(-1).squeeze(-1).squeeze(-1)
# Quantification predictions
diameters = self.diameter_regressor(pooled_features)
angles = self.angle_regressor(pooled_features)
return anterior_pred, posterior_pred, diameters, angles
def postprocess(self, volume, segmentation, anterior_class, posterior_class):
"""
Post-processing pipeline for anatomical quantification
Based on Snaillab's eICAB approach with enhancements
"""
# Convert to numpy for post-processing
seg_np = segmentation.squeeze().cpu().numpy()
vol_np = volume.squeeze().cpu().numpy()
# 1. Centerline extraction
skeleton = skeletonize(seg_np > 0)
# 2. Bifurcation detection
bifurcations = self.detect_bifurcations(skeleton)
# 3. Artery diameter measurement
diameters = self.measure_diameters(vol_np, skeleton, bifurcations)
# 4. Bifurcation angle calculation
angles = self.calculate_angles(skeleton, bifurcations)
return diameters, angles
def detect_bifurcations(self, skeleton):
"""
Detect bifurcation points in skeletonized vessels
"""
from scipy.ndimage import convolve
kernel = np.ones((3, 3, 3))
conv = convolve(skeleton.astype(int), kernel, mode='constant')
# Bifurcation points have 3 or more neighbors
bifurcations = (conv >= 4) & skeleton
return np.argwhere(bifurcations)
def measure_diameters(self, volume, skeleton, bifurcations, distance=5):
"""
Measure artery diameters at specified locations
"""
# Distance transform to vessel boundaries
dist_map = distance_transform_edt(skeleton)
diameters = {}
artery_types = ['A1', 'A2', 'M1', 'M2', 'ICA', 'PcoA', 'P1', 'BA', 'VA']
for artery in artery_types:
# For simplicity - actual implementation would use anatomical knowledge
# to locate specific measurement points
if artery in ['A2', 'P1', 'ICA', 'VA']:
# Measure 5mm beyond bifurcation
points = self.locate_measurement_points(artery, bifurcations, skeleton, distance)
else:
# Measure at 50% of artery length
points = self.mid_artery_points(artery, skeleton)
# Calculate diameter as twice the distance to boundary
diam_values = 2 * dist_map[tuple(points.T)]
diameters[artery] = np.mean(diam_values)
return diameters
def calculate_angles(self, skeleton, bifurcations):
"""
Calculate bifurcation angles using vector geometry
"""
angles = {}
vectors = {}
for i, point in enumerate(bifurcations):
# Get connected branches
branches = self.get_branches(point, skeleton)
if len(branches) < 3:
continue
# Create vectors from bifurcation point
vecs = [branch - point for branch in branches[:3]]
# Calculate angles between vectors
angle1 = self.vector_angle(vecs[0], vecs[1])
angle2 = self.vector_angle(vecs[1], vecs[2])
angle3 = self.vector_angle(vecs[0], vecs[2])
angles[f'bifurcation_{i}'] = [angle1, angle2, angle3]
return angles
def vector_angle(self, v1, v2):
"""Calculate angle between two vectors in degrees"""
cos_theta = np.dot(v1, v2) / (np.linalg.norm(v1) * np.linalg.norm(v2))
return np.degrees(np.arccos(np.clip(cos_theta, -1.0, 1.0)))
# ------------ Utility Functions for Data Processing ------------
def load_and_preprocess_mra(file_path):
"""
Load and preprocess TOF-MRA NIfTI file
"""
img = nib.load(file_path)
data = img.get_fdata()
affine = img.affine
# 1. Intensity normalization
data = (data - np.min(data)) / (np.max(data) - np.min(data))
# 2. Skull stripping (simplified)
mask = data > 0.1
data = data * mask
# 3. Resampling to isotropic resolution (1mm³)
# (Actual implementation would use interpolation)
# 4. Zero-centering
data = (data - 0.5) * 2.0
return torch.tensor(data).float().unsqueeze(0).unsqueeze(0)
def load_annotations(annotation_path):
"""
Load CROWN Challenge annotations from JSON
"""
with open(annotation_path) as f:
data = json.load(f)
return data
# ------------ Training and Evaluation ------------
def train(model, dataloader, optimizer, device):
model.train()
total_loss = 0.0
for volume, anterior_label, posterior_label, diameters, angles in dataloader:
volume = volume.to(device)
anterior_label = anterior_label.to(device)
posterior_label = posterior_label.to(device)
diameters = diameters.to(device)
angles = angles.to(device)
optimizer.zero_grad()
# Forward pass
ant_pred, post_pred, diam_pred, angle_pred = model(volume)
# Calculate losses
class_loss = F.cross_entropy(ant_pred, anterior_label) + \
F.cross_entropy(post_pred, posterior_label)
quant_loss = F.mse_loss(diam_pred, diameters) + \
F.mse_loss(angle_pred, angles)
loss = class_loss + quant_loss
loss.backward()
optimizer.step()
total_loss += loss.item()
return total_loss / len(dataloader)
def evaluate(model, dataloader, device):
model.eval()
metrics = {
'anterior_acc': 0,
'posterior_acc': 0,
'diam_mae': 0,
'angle_mae': 0
}
with torch.no_grad():
for volume, ant_label, post_label, diameters, angles in dataloader:
volume = volume.to(device)
# Forward pass
ant_pred, post_pred, diam_pred, angle_pred = model(volume)
# Classification accuracy
_, ant_pred_class = torch.max(ant_pred, 1)
_, post_pred_class = torch.max(post_pred, 1)
metrics['anterior_acc'] += (ant_pred_class == ant_label).sum().item()
metrics['posterior_acc'] += (post_pred_class == post_label).sum().item()
# Quantification MAE
metrics['diam_mae'] += F.l1_loss(diam_pred, diameters).item()
metrics['angle_mae'] += F.l1_loss(angle_pred, angles).item()
# Normalize metrics
n = len(dataloader.dataset)
metrics['anterior_acc'] /= n
metrics['posterior_acc'] /= n
metrics['diam_mae'] /= len(dataloader)
metrics['angle_mae'] /= len(dataloader)
return metrics
# ------------ Main Workflow ------------
if __name__ == "__main__":
# Configuration
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
num_epochs = 50
lr = 1e-4
# Initialize model
model = CoWModel().to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
# Load dataset (placeholder)
# In practice, use CROWN challenge dataset from:
# https://doi.org/10.34894/R05G1L
train_loader = None # Replace with actual DataLoader
val_loader = None # Replace with actual DataLoader
# Training loop
for epoch in range(num_epochs):
train_loss = train(model, train_loader, optimizer, device)
val_metrics = evaluate(model, val_loader, device)
print(f"Epoch {epoch+1}/{num_epochs}")
print(f"Train Loss: {train_loss:.4f}")
print(f"Validation - Ant Acc: {val_metrics['anterior_acc']:.4f}, "
f"Post Acc: {val_metrics['posterior_acc']:.4f}, "
f"Diam MAE: {val_metrics['diam_mae']:.4f}, "
f"Angle MAE: {val_metrics['angle_mae']:.4f}")
# Save model
torch.save(model.state_dict(), "crown_model.pth")# Usage Instructions
# Load TOF-MRA volume
volume = load_and_preprocess_mra("path/to/tof_mra.nii.gz")
# Load annotations
annotations = load_annotations("path/to/annotations.json")
# Initialize model
model = CoWModel().to(device)
# Train for 50 epochs
for epoch in range(50):
train_loss = train(model, train_loader, optimizer, device)
# Load pretrained model
model.load_state_dict(torch.load("crown_model.pth"))
# Forward pass
anterior_pred, posterior_pred, diameters, angles = model(volume)
# Post-processing for anatomical quantification
final_diameters, final_angles = model.postprocess(
volume,
segmentation,
anterior_pred,
posterior_pred
)
metrics = evaluate(model, test_loader, device)
print(f"Anterior Accuracy: {metrics['anterior_acc']:.4f}")
print(f"Diameter MAE: {metrics['diam_mae']:.4f} mm")
# Use mixed precision training
scaler = torch.cuda.amp.GradScaler()
with torch.cuda.amp.autocast():
outputs = model(inputs)
loss = criterion(outputs, targets)
scaler.scale(loss).backward()
scaler.step(optimizer)
scaler.update()
# Add to data loader
transform = Compose([
RandRotate(range_x=15, prob=0.5),
RandZoom(prob=0.5, min_zoom=0.9, max_zoom=1.1),
RandGaussianNoise(prob=0.5, std=0.01),
RandAdjustContrast(prob=0.5, gamma=(0.7, 1.3))
])
# Use gradient checkpointing
model.segmentor = torch.utils.checkpoint(model.segmentor)
# Prune redundant filters
prune.l1_unstructured(model.anterior_classifier.fc, name='weight', amount=0.2)FAQs: Everything You Need to Know About Circle of Willis Imaging
Q: What is the Circle of Willis?
A: It’s a ring-like arterial network at the base of the brain that ensures continuous blood supply to cerebral regions.
Q: Why is it important in stroke prevention?
A: Variations in the CoW, such as narrow arteries or abnormal bifurcations, are associated with higher risks of intracranial aneurysms and ASAH.
Q: How does AI help in analyzing the Circle of Willis?
A: AI automates classification of anatomical variants and measurement of artery diameters and angles, improving speed and consistency.
Q: What is the CROWN challenge?
A: It’s a scientific competition designed to benchmark AI techniques for automated CoW classification and quantification using TOF-MRA scans.
Q: Are these AI tools ready for clinical use?
A: Not yet — while promising, current models still lack the precision required for standalone clinical decision-making.
References
Vos, I.N. et al. (2025). Medical Image Analysis, 105, 103650. DOI
Yang, K. et al. (2023). TopCoW Challenge: GitHub
Metrics Reloaded Framework: Nature Methods

Pingback: ElastoNet 1: The Revolutionary Neural Network for MRE Wave Inversion with Uncertainty Quantification (Pros & Cons) - aitrendblend.com