The Crippling Burden of Breast Cancer Radiotherapy Planning (And the AI Solution Changing Everything)
Every 38 seconds, a woman is diagnosed with breast cancer globally. For these patients, timely and precise radiotherapy is often a lifeline. Yet, the complex, multi-step process of planning this treatment – involving synthesizing medical reports, defining treatment strategies, and meticulously mapping 3D target volumes – is notoriously slow, error-prone, and burns out clinical teams. Radiation oncologists spend hours per patient on tasks ripe for automation, while subtle inconsistencies between steps can lead to under-dosing the tumor or over-exposing healthy tissue.
This is the problem RO-LMM solves. Groundbreaking research published in Medical Image Analysis (Kim et al., 2025) introduces RO-LMM (Radiation Oncology Large Multimodal Model), the world’s first end-to-end AI assistant specifically designed for the radiation oncology workflow. RO-LMM isn’t just another single-task algorithm; it’s a comprehensive foundation model that tackles the entire sequence of critical planning tasks with unprecedented speed and accuracy. Forget incremental improvements – RO-LMM represents a paradigm shift, leveraging advanced “Consistency Embedding” techniques (CEFTune & CESEG) to eliminate dangerous error accumulation and deliver results that generalize across hospitals. The results? Up to 20% higher accuracy in complex cases and planning completed in under 10 seconds per task.
Why Traditional Methods Are Failing Breast Cancer Patients (The Pain Points RO-LMM Obliterates)
Current radiotherapy planning is a fragmented, high-pressure bottleneck:
- The Information Overload Nightmare: Oncologists drown in dense MRI, ultrasound, and pathology reports. Manually distilling this into a concise clinical summary is time-consuming and subjective. (Keyword: clinical report summarization)
- The Strategy Guessing Game: Translating that summary into an optimal radiotherapy plan (dose, technique, target areas) relies heavily on individual expertise and institutional protocols, leading to variability. (Keyword: radiotherapy strategy suggestion)
- The Delicate Art (and Error) of Delineation: Manually drawing the precise 3D “Clinical Target Volume” (CTV) – the area receiving radiation – on CT scans is painstakingly slow (30-60+ minutes) and highly susceptible to inter-observer variability. A slight miscalculation here can mean missing cancer cells or damaging the heart or lungs. (Keyword: 3D target volume segmentation)
- The Domino Effect of Errors: Crucially, mistakes in step 1 or 2 cascade catastrophically into step 3. If the summary misses a key detail or the strategy is ambiguous, the segmentation will be wrong, no matter how good the segmentation tool is. Traditional AI tools, focused on single tasks, utterly fail here. (Keyword: error accumulation in radiotherapy)
- Workload Burnout: This labor-intensive process contributes significantly to clinician burnout and treatment delays.
RO-LMM: Your AI Clinical Partner for End-to-End Radiotherapy Excellence (How It Works)
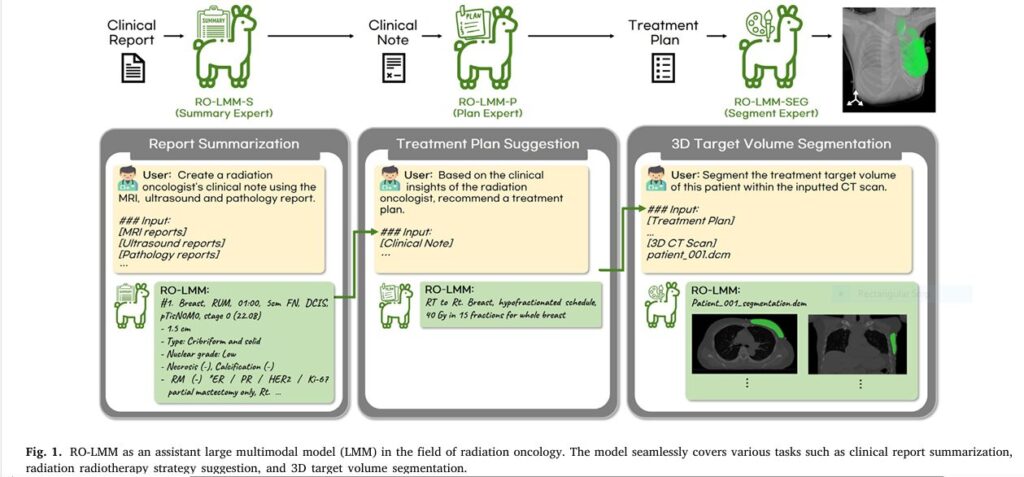
RO-LMM isn’t just automation; it’s intelligent, multimodal collaboration. Think of it as an expert clinical assistant powered by cutting-edge AI foundation models, specifically fine-tuned for the nuances of radiation oncology. Here’s how it transforms the workflow (Fig. 1):
- RO-LMM-S (The Summary Expert):
- Input: Raw MRI, Ultrasound, and Pathology reports.
- Action: Instantly generates a concise, accurate, and clinically relevant summary note – just like an experienced oncologist would. No more report digging.
- Keyword Integration: Clinical context summarization, multimodal data fusion.
- RO-LMM-P++ (The Plan Expert – Enhanced):
- Input: The AI-generated clinical summary note.
- Action: Proposes an optimal, personalized radiotherapy strategy (e.g., “RT to Rt. Breast, Hypofractionated, WBI with SIB boost”). It leverages CEFTune (Consistency Embedding Fine-Tuning) – the secret weapon. CEFTune injects controlled noise during training and enforces consistency between outputs from clean and noisy inputs. This makes RO-LMM-P++ robust to imperfections in the summary note, preventing early errors from derailing the whole process.
- Keyword Integration: Treatment plan suggestion, AI clinical decision support, CEFTune, robust AI.
- RO-LMM-SEG++ (The Segment Expert – Enhanced):
- Input: The AI-generated treatment plan + The patient’s 3D CT scan.
- Action: Automatically segments the complex 3D target volume (breast, chest wall, lymph nodes) directly on the CT scan, perfectly aligned with the proposed strategy. This is where CESEG (Consistency Embedding Segmentation) shines. CESEG ensures the segmentation model performs flawlessly whether it receives a perfectly clean plan or one generated by RO-LMM-P++, maintaining spatial accuracy and eliminating error propagation from previous steps.
- Keyword Integration: Plan-guided segmentation, AI contouring, CESEG, 3D medical image segmentation, error propagation prevention.
The Revolutionary Tech Inside: CEFTune & CESEG
The true genius of RO-LMM lies in its novel training techniques designed to combat the fatal flaw of sequential AI tasks: error accumulation.
- CEFTune: Trains the “Plan Expert” (RO-LMM-P++) to be incredibly resilient. By adding noise to input embeddings during training and enforcing that the model’s outputs remain consistent whether the input is clean or noisy, CEFTune ensures reliable strategy generation even if the initial summary has minor flaws. It uses SentenceBERT to measure semantic consistency.
- CESEG: Extends this consistency principle to the complex 3D segmentation task. It trains the “Segment Expert” (RO-LMM-SEG++) to produce near-identical, accurate segmentations whether it receives a pristine ground-truth plan or the actual plan generated by RO-LMM-P++ during the workflow. The performance drop vanishes (Table 9).
Proven Results: RO-LMM Outperforms Humans & Existing AI (The Data Doesn’t Lie)
This isn’t theoretical. Rigorous multi-center validation (Yonsei Cancer Center & Yongin Severance Hospital) on real patient data proves RO-LMM’s superiority:
- Clinical Report Summarization (RO-LMM-S):
- Crushed ChatGPT & Medical LLMs: Achieved Rouge-L scores of 0.788 (Internal) and 0.788 (External) vs. ChatGPT’s 0.685 and 0.683 (Table 2).
- Clinical Experts Preferred It: Scored significantly higher (45/50 vs 13.3/50) on expert rubrics for relevance, accuracy, and conciseness (Table 3). Clinicians agreed strongly (correlation >0.85).
- Keyword Validation: AI report summarization accuracy, outperforming ChatGPT.
- Radiotherapy Strategy Suggestion (RO-LMM-P++):
- Destroyed GPT-4.0 & Specialized Models: Scored Rouge-L 0.655 (Internal) and 0.615 (External), far exceeding GPT-4.0’s 0.356 and 0.316 (Table 2). On public data, it scored 0.669 vs. GPT-4’s 0.390 (Table 7).
- Clinically Superior: Experts rated RO-LMM-P++ plans highest (42.8/50 vs GPT-4’s 30.8/50), especially for correctly defining complex fields (R3) and dose/fractionation (R4) (Table 4). CEFTune was key to this external validation success.
- Keyword Validation: AI treatment planning accuracy, beating GPT-4, domain-specific AI.
- 3D Target Volume Segmentation (RO-LMM-SEG++):
- Massive Gains Over Unimodal AI: Achieved Dice scores of 0.828 (Internal) and 0.761 (External) vs. standard 3D U-Net’s 0.782 and 0.700 (Table 5). HD95 (accuracy measure) plummeted from 45.8mm to 12.6mm internally.
- Handled Complexity Flawlessly: For atypical cases (e.g., post-mastectomy), gains were even more dramatic – up to 20% Dice improvement externally (Table 6).
- CESEG Eliminated the Noise Gap: Performance using AI-generated plans vs. perfect ground-truth plans differed by less than 1% (Table 9). Traditional models showed significant drops (>5%).
- Visually Perfect: Qualitative results (Fig. 4) show RO-LMM-SEG++ accurately contouring complex breast/lymph node regions, while unimodal AI failed spectacularly.
- Keyword Validation: AI contouring accuracy, automatic target delineation, CESEG benefits, Dice score improvement.
- Speed & Efficiency: Completes the entire sequence of tasks per patient in ~10 seconds using a single NVIDIA A6000 GPU (24GB RAM) – orders of magnitude faster than manual planning (Table 11). Enables local, privacy-preserving deployment.
- Keyword Validation: Faster radiotherapy planning, efficient AI clinical workflow.
If you’re Interested in advance methods in medical imaging, you may also find this article helpful: 3 Breakthroughs & 1 Warning: How Explainable AI SVIS-RULEX is Revolutionizing Medical Imaging (Finally!)
Beyond the Hype: Addressing Limitations & The Road Ahead
RO-LMM is revolutionary, but research continues:
- Current Scope: Focused on initial breast cancer cases (post-BCS or mastectomy). Boost target delineation (e.g., tumor bed) requires integrating surgical clips/seromas from multiple scans – a future challenge.
- Prescription Variability: Training data included institutional practices. Future versions need stricter standardization on evidence-based dosing.
- EMR Integration: Token limits restricted inputs to core reports (MRI, US, Path). Expanding to full EMR data is crucial.
- Generalization: Proven across two Korean centers. Validation in diverse global populations and healthcare systems is ongoing. Early public dataset tests (Table 7) are highly promising.
The Future is Multimodal & Integrated: RO-LMM paves the way for true “generalist medical AI.” Imagine this model expanding to lung, prostate, or brain cancer, incorporating genomics, and seamlessly integrating with hospital EMR and treatment machines. The potential to democratize high-quality cancer care globally is immense.
The Imperative Call to Action: Don’t Let Your Patients Wait
The era of slow, error-prone, manual radiotherapy planning is ending. RO-LMM demonstrates that AI can handle complex clinical workflows end-to-end with superhuman speed and accuracy, significantly reducing the risk of dangerous errors and freeing oncologists for patient care.
Here’s What You Can Do Right Now:
- Oncologists & Clinics: Demand AI-powered planning tools. Ask vendors about integrated solutions leveraging multimodal LMMs like RO-LMM. Prioritize systems with consistency techniques (CEFTune/CESEG) to ensure safety. Explore pilot programs.
- Researchers: Dive into the paper. Replicate the methods. Investigate extending RO-LMM to other cancers and integrating more data modalities (genomics, PET). Contribute to open-source efforts or synthetic datasets like the one provided by the authors.
- Hospital Administrators: Invest in this technology. Calculate the ROI: reduced planning time, minimized errors (and costly re-plans), improved patient throughput, enhanced staff satisfaction, and superior patient outcomes. This is not just tech; it’s a strategic advantage.
- Patients: Ask your care team: “Are you using the latest AI-assisted planning techniques to ensure the fastest, most accurate treatment possible for my breast cancer?”
The future of precise, accessible, and efficient radiotherapy is here. Embrace RO-LMM and be part of the revolution in cancer care. Let’s move beyond human limitations and deliver the best possible outcomes, faster.
Here’s the complete PyTorch implementation of the RO-LMM framework with CEFTune and CESEG techniques:
import torch
import torch.nn as nn
from transformers import LlamaForCausalLM, LlamaTokenizer, LlamaModel
from sentence_transformers import SentenceTransformer
from monai.networks.nets import UNet
class ConsistencyEmbeddingFineTuning(nn.Module):
"""
Implements CEFTune for radiotherapy strategy suggestion (RO-LMM-P++)
"""
def __init__(self, base_model_name="Llama-2-7b-chat"):
super().__init__()
self.llama = LlamaForCausalLM.from_pretrained(base_model_name)
self.tokenizer = LlamaTokenizer.from_pretrained(base_model_name)
self.sbert = SentenceTransformer('NeuML/pubmedbert-base-embeddings')
self.alpha = 5 # Noise scaling factor
self.lambda_consistency = 1.0 # Consistency loss weight
def add_noise_to_embeddings(self, embeddings):
"""Add uniform noise to embeddings (NEFTune technique)"""
L, C = embeddings.shape[1], embeddings.shape[2]
noise = torch.rand_like(embeddings) * 2 - 1 # Uniform [-1, 1]
return embeddings + (self.alpha / (L * C)**0.5) * noise
def forward(self, input_ids, attention_mask, labels=None):
# Get clean embeddings
embeddings = self.llama.model.embed_tokens(input_ids)
# Create noisy embeddings
noisy_embeddings = self.add_noise_to_embeddings(embeddings)
# Process clean inputs
outputs_clean = self.llama(
inputs_embeds=embeddings,
attention_mask=attention_mask,
labels=labels
)
# Process noisy inputs
outputs_noisy = self.llama(
inputs_embeds=noisy_embeddings,
attention_mask=attention_mask,
labels=labels
)
# Calculate cross-entropy loss
ce_loss = outputs_noisy.loss
# Calculate consistency loss
clean_logits = outputs_clean.logits
noisy_logits = outputs_noisy.logits
# Detokenize and get sentence embeddings
clean_sentences = [self.tokenizer.decode(ids, skip_special_tokens=True)
for ids in clean_logits.argmax(-1)]
noisy_sentences = [self.tokenizer.decode(ids, skip_special_tokens=True)
for ids in noisy_logits.argmax(-1)]
clean_emb = self.sbert.encode(clean_sentences, convert_to_tensor=True)
noisy_emb = self.sbert.encode(noisy_sentences, convert_to_tensor=True)
# Cosine similarity loss
consistency_loss = 1 - torch.cosine_similarity(clean_emb, noisy_emb).mean()
# Total loss
total_loss = ce_loss + self.lambda_consistency * consistency_loss
return {
"loss": total_loss,
"logits": outputs_clean.logits,
"ce_loss": ce_loss,
"consistency_loss": consistency_loss
}
class RO_LMM_SEG(nn.Module):
"""
Implements CESEG for target volume segmentation (RO-LMM-SEG++)
"""
def __init__(self, text_model_path, img_size=(384, 384, 128)):
super().__init__()
# Frozen LLM for text processing
self.llama = LlamaModel.from_pretrained(text_model_path)
for param in self.llama.parameters():
param.requires_grad = False
# Learnable text prompts
self.prompt_embeddings = nn.Parameter(torch.randn(10, self.llama.config.hidden_size))
# Image segmentation backbone
self.unet = UNet(
spatial_dims=3,
in_channels=1,
out_channels=1,
channels=(16, 32, 64, 128, 256),
strides=(2, 2, 2, 2),
num_res_units=2
)
# Multimodal fusion layers
self.fusion_conv = nn.Conv3d(256 + 128, 256, kernel_size=1)
self.attention = nn.MultiheadAttention(256, num_heads=8, batch_first=True)
# CESEG parameters
self.alpha = 5
self.lambda_consistency = 1.0
def process_text(self, input_ids, attention_mask, noise=False):
# Get text embeddings
embeddings = self.llama.embed_tokens(input_ids)
# Add noise if requested (for CESEG)
if noise:
L, C = embeddings.shape[1], embeddings.shape[2]
noise_tensor = torch.rand_like(embeddings) * 2 - 1
embeddings = embeddings + (self.alpha / (L * C)**0.5) * noise_tensor
# Add learnable prompts
prompt_embeds = self.prompt_embeddings.unsqueeze(0).repeat(embeddings.shape[0], 1, 1)
embeddings = torch.cat([prompt_embeds, embeddings], dim=1)
# Process through LLM
outputs = self.llama(inputs_embeds=embeddings, attention_mask=attention_mask)
text_emb = outputs.last_hidden_state[:, 0] # CLS token
return text_emb
def forward(self, ct_volume, text_inputs, clean_text=None):
# Unpack text inputs
input_ids, attention_mask = text_inputs
# Process image
img_features = self.unet(ct_volume)
# Process text (with noise)
text_emb_noisy = self.process_text(input_ids, attention_mask, noise=True)
# Prepare text embedding for fusion
text_emb_noisy = text_emb_noisy.view(-1, 128, 1, 1, 1)
text_emb_noisy = text_emb_noisy.expand(-1, -1, *img_features.shape[2:])
# Multimodal fusion
fused_features = torch.cat([img_features, text_emb_noisy], dim=1)
fused_features = self.fusion_conv(fused_features)
# Attention refinement
B, C, D, H, W = fused_features.shape
features_flat = fused_features.view(B, C, -1).permute(0, 2, 1)
attn_output, _ = self.attention(features_flat, features_flat, features_flat)
attn_output = attn_output.permute(0, 2, 1).view(B, C, D, H, W)
# Segmentation prediction
seg_pred = torch.sigmoid(attn_output)
# CESEG consistency branch
if self.training and clean_text is not None:
# Process clean text
clean_input_ids, clean_attention_mask = clean_text
text_emb_clean = self.process_text(clean_input_ids, clean_attention_mask, noise=False)
# Calculate consistency loss
consistency_loss = 1 - torch.cosine_similarity(
text_emb_noisy.view(B, -1),
text_emb_clean.view(B, -1)
).mean()
return seg_pred, consistency_loss
return seg_pred
class RO_LMM_System:
"""
End-to-end RO-LMM system for breast cancer radiotherapy planning
"""
def __init__(self, device='cuda'):
self.device = device
# Initialize components
self.summary_expert = LlamaForCausalLM.from_pretrained(
"path/to/fine-tuned/RO-LMM-S"
).to(device)
self.plan_expert = ConsistencyEmbeddingFineTuning(
base_model_name="path/to/fine-tuned/RO-LMM-P-base"
).to(device)
self.segment_expert = RO_LMM_SEG(
text_model_path="path/to/RO-LMM-P++"
).to(device)
# Tokenizers
self.tokenizer = LlamaTokenizer.from_pretrained("Llama-2-7b-chat")
self.tokenizer.add_special_tokens({'pad_token': '[PAD]'})
def generate_clinical_summary(self, mri_report, us_report, pathology_report):
"""
RO-LMM-S: Generate clinical note from multimodal reports
"""
prompt = f"""
Create a radiation oncologist's clinical note using these reports:
MRI Report: {mri_report}
Ultrasound Report: {us_report}
Pathology Report: {pathology_report}
"""
inputs = self.tokenizer(
prompt,
return_tensors="pt",
max_length=4096,
truncation=True
).to(self.device)
outputs = self.summary_expert.generate(
**inputs,
max_new_tokens=256,
temperature=0.7,
top_p=0.9
)
summary = self.tokenizer.decode(outputs[0], skip_special_tokens=True)
return summary.split("Clinical Note:")[-1].strip()
def suggest_radiotherapy_plan(self, clinical_summary):
"""
RO-LMM-P++: Generate radiotherapy strategy from clinical summary
"""
prompt = f"""
Based on this clinical summary, recommend a radiotherapy plan:
{clinical_summary}
"""
inputs = self.tokenizer(
prompt,
return_tensors="pt",
max_length=4096,
truncation=True,
padding=True
).to(self.device)
with torch.no_grad():
outputs = self.plan_expert(**inputs)
logits = outputs['logits']
plan = self.tokenizer.decode(
logits.argmax(-1)[0],
skip_special_tokens=True
)
return plan.split("Radiotherapy Plan:")[-1].strip()
def segment_target_volume(self, ct_volume, radiotherapy_plan):
"""
RO-LMM-SEG++: Perform 3D segmentation using CT and radiotherapy plan
"""
# Preprocess CT volume
ct_volume = self._preprocess_ct(ct_volume).to(self.device)
# Tokenize radiotherapy plan
text_inputs = self.tokenizer(
radiotherapy_plan,
return_tensors="pt",
max_length=512,
truncation=True,
padding='max_length'
)
text_inputs = (text_inputs['input_ids'].to(self.device),
text_inputs['attention_mask'].to(self.device))
# Perform segmentation
with torch.no_grad():
segmentation = self.segment_expert(
ct_volume.unsqueeze(0), # Add batch dim
text_inputs
)
return segmentation.squeeze(0).cpu().numpy()
def end_to_end_planning(self, mri, us, pathology, ct_volume):
"""Complete end-to-end radiotherapy planning workflow"""
print("Generating clinical summary...")
summary = self.generate_clinical_summary(mri, us, pathology)
print("\nSuggesting radiotherapy plan...")
plan = self.suggest_radiotherapy_plan(summary)
print("\nSegmenting target volume...")
segmentation = self.segment_target_volume(ct_volume, plan)
return {
"clinical_summary": summary,
"radiotherapy_plan": plan,
"segmentation_mask": segmentation
}
def _preprocess_ct(self, ct_volume):
"""Preprocess CT volume (truncation and normalization)"""
# Truncate HU values (-1000 to 1000)
ct_volume = torch.clamp(ct_volume, -1000, 1000)
# Normalize to [0, 1]
ct_volume = (ct_volume + 1000) / 2000
# Resample to (384, 384, 128) if needed
if ct_volume.shape != (384, 384, 128):
ct_volume = nn.functional.interpolate(
ct_volume.unsqueeze(0).unsqueeze(0),
size=(128, 384, 384),
mode='trilinear'
).squeeze()
return ct_volume
# Example Usage
if __name__ == "__main__":
# Initialize system (requires pre-trained weights)
ro_lmm = RO_LMM_System()
# Example inputs (in real usage, load actual medical data)
mri_report = "Right breast mass at 2 o'clock, size 1.5 cm, BIRADS 4"
us_report = "Hypoechoic irregular mass, size 1.6 cm, suspicious for malignancy"
pathology_report = "Invasive ductal carcinoma, ER+, PR+, HER2-"
ct_volume = torch.randn(512, 512, 120) # Simulated CT data
# Run end-to-end planning
results = ro_lmm.end_to_end_planning(
mri_report, us_report, pathology_report, ct_volume
)
print("\nResults:")
print(f"Clinical Summary: {results['clinical_summary']}")
print(f"Radiotherapy Plan: {results['radiotherapy_plan']}")
print(f"Segmentation Mask Shape: {results['segmentation_mask'].shape}")Sources & Further Reading:
- Kim, K., Oh, Y., Park, S., et al. (2025). End-to-end breast cancer radiotherapy planning via LMMs with consistency embedding. Medical Image Analysis, 105, 103646. https://doi.org/10.1016/j.media.2025.103646
- Oh, Y., Park, S., Byun, H.K., et al. (2024). LLM-driven multimodal target volume contouring in radiation oncology. Nature Communications, 15(1), 9186.
- Jain, N., et al. (2024). NEFTune: Noisy Embeddings Improve Instruction Finetuning. ICLR 2024.

Pingback: Title: 5 Powerful Reasons Why Counterfactual Contrastive Learning Beats Traditional Medical Imaging Techniques (And How It Can Transform Your Practice) - aitrendblend.com