New Deep Learning Model Boosts Accuracy for Early Detection
Skin cancer, particularly melanoma, remains one of the deadliest cancers worldwide. The stakes for early detection couldn’t be higher: diagnose melanoma at an advanced stage, and the 10-year survival rate plummets to a grim 39%. Catch it early, however, and survival rates soar above 93%. This life-or-death difference underscores the critical need for accurate, accessible diagnostic tools.
Enter artificial intelligence (AI). Groundbreaking research from Atatürk University, published in Engineering Applications of Artificial Intelligence, introduces a novel deep learning framework that significantly improves multi-label skin lesion classification. This AI system, leveraging multiple data sources and sophisticated attention mechanisms, achieved a remarkable 83.04% average accuracy – outperforming the best existing methods by over 2%. Let’s explore how this innovation is transforming dermatology.
Keywords: skin cancer AI diagnosis, multi-label lesion classification, dermoscopy deep learning, melanoma detection AI, computer-aided diagnosis skin, seven-point checklist AI, hybrid deep learning dermatology.
The Diagnostic Challenge: Beyond the Naked Eye
Diagnosing skin cancer is complex. Dermatologists traditionally rely on:
- Clinical Images: Captured with standard digital cameras, showing global features like shape, color, borders, and elevation. However, they lack subsurface detail and suffer from inconsistencies in lighting and framing.
- Dermoscopy Images: Taken with specialized tools (using liquid immersion or polarized light) to minimize surface reflection. These provide magnified (10x) views revealing crucial subsurface structures like pigment networks, dots, globules, and vascular patterns. Dermoscopy significantly boosts diagnostic accuracy but requires expertise.
- Patient Meta-Data: Age, sex, medical history, sun exposure, and family history provide essential context for risk assessment.
Adding to the complexity is the “Seven-Point Checklist” (SPC), a standardized diagnostic tool evaluating seven visual criteria (Pigment Network, Blue-Whitish Veil, Vascular Structures, etc.). Each criterion is scored (major=2 points, minor=1 point), with a total score ≥ 3 suggesting melanoma. Simultaneously evaluating these multiple criteria across different image types is inherently subjective and challenging.
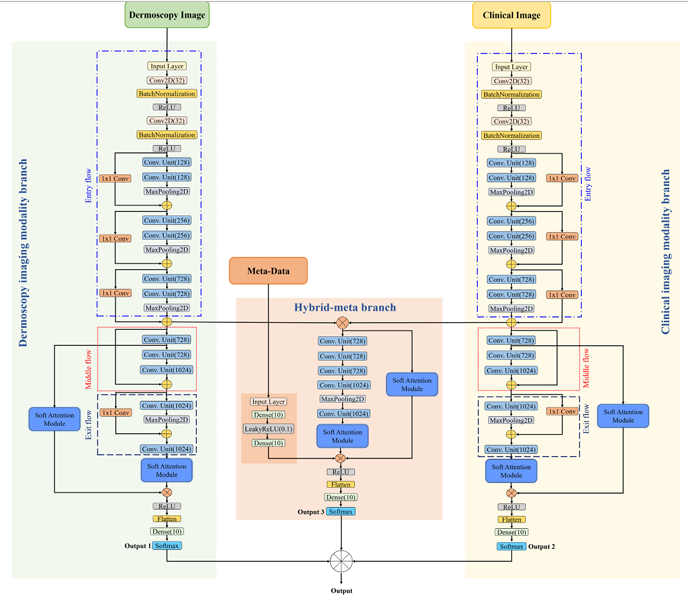
Introducing the Hybrid Soft Attention Multi-modal Framework
The Turkish research team tackled these challenges head-on with a sophisticated deep learning architecture designed to mimic and enhance the dermatologist’s multi-faceted approach:
- A Three-Branch Architecture:
- Dermoscopy Imaging Branch: Processes high-magnification dermoscopy images using a modified Xception network backbone.
- Clinical Imaging Branch: Processes broader-view clinical photographs, also using a modified Xception network.
- Hybrid-Meta Branch: The innovation hub. It fuses the feature maps extracted from the Dermoscopy and Clinical branches. Crucially, it then concatenates these fused image features with the patient’s meta-data (age, sex, etc.). This branch employs multiple convolutional units to learn the complex relationships between the fused image data and the contextual patient information.
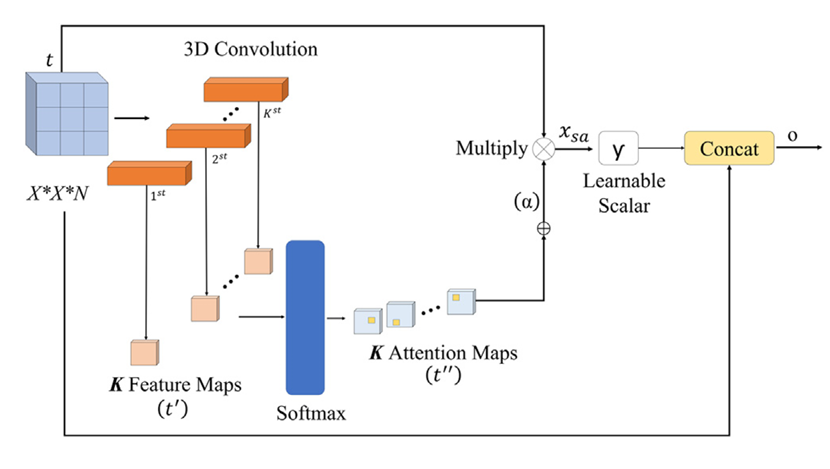
- The Power of Soft Attention:
Not all pixels are created equal. Skin lesions often occupy a small portion of an image, surrounded by irrelevant features like hair, veins, or skin texture. The researchers integrated Soft Attention Modules within each branch. These modules act like a dynamic spotlight:- Analyze the feature maps generated by the network.
- Identify and assign higher importance (weights) to regions most relevant for diagnosing the lesion.
- Generate “attention maps” that highlight these critical areas.
- Enhance the discriminative power of the features by focusing computations on salient regions.
Result: The network learns to “pay attention” to the medically significant parts of the image, just like an expert dermatologist would, significantly boosting feature quality.
- Intelligent Hybrid Fusion:
Instead of making decisions based on a single branch, the framework employs a late fusion strategy:- Each branch (Dermoscopy, Clinical, Hybrid-Meta) independently processes its specific input and produces probability scores for each possible label (the 7 SPC criteria + the final diagnosis).
- The final prediction for each label is determined by averaging the probability scores from all three branches.
- The label with the highest average probability is chosen.
This leverages the complementary strengths of each data modality – the microscopic detail of dermoscopy, the macroscopic context of clinical images, and the patient-specific context of meta-data – leading to more robust and accurate predictions.
Why Modified Xception?
The Xception network was chosen as the backbone for the image branches due to its efficiency and performance. Its key innovation is depthwise separable convolutions, which dramatically reduce computational cost while maintaining or even improving accuracy compared to predecessors like Inception V3. The researchers further optimized it for medical imaging by strategically reducing redundant layers in its middle flow, making it faster to train on smaller medical datasets without sacrificing performance.
Performance That Sets a New Standard
The model was rigorously tested on the publicly available Seven-Point Criteria Evaluation (SPC) dataset, comprising data from 1,011 patients (dermoscopy images, clinical images, meta-data, SPC labels, and diagnosis). Results were compelling:
- State-of-the-Art Accuracy: Achieved an Average Accuracy (AVG) of 83.04% across all seven SPC criteria and the final diagnosis label.
- Significant Improvement: Outperformed the previous best method (Constrained Classifier Chain – CC) by 2.14% in AVG. This represents a major leap in a field where incremental gains are hard-won.
- Attention Matters: An ablation study proved the critical role of the Soft Attention Module. Adding attention boosted the average accuracy by 4.2% (from 79.68% to 83.04%).
- Multi-modal Advantage: The hybrid fusion of dermoscopy, clinical images, and meta-data consistently outperformed models using only one or two modalities. For example:
- Dermoscopy alone: 79.72% AVG
- Clinical alone: 70.57% AVG
- Dermoscopy + Clinical: 81.23% AVG
- Dermoscopy + Clinical + Meta-data (Proposed): 83.04% AVG
- Superior AUC: The proposed model achieved the highest Area Under the Curve (AUC) scores for all label types compared to leading methods like Inception-Combined and FusionM4Net-SS, indicating excellent overall discriminative power.
- Robustness: Achieved high performance despite significant class imbalance in the SPC dataset – a common and challenging real-world problem.
Table: Performance Comparison Against State-of-the-Art Methods (Accuracy %)
| Method | BWV | DAG | PIG | PN | RS | STR | VS | DIAG | AVG |
|---|---|---|---|---|---|---|---|---|---|
| Alzahrani et al. (2019) | 80.5 | 50.4 | 62.8 | 62.8 | 71.9 | 67.6 | 73.2 | 64.3 | 66.7 |
| EmbeddingNet (Yap et al.) | 84.3 | 57.5 | 64.3 | 65.1 | 78.0 | 73.4 | 82.5 | 68.6 | 71.7 |
| Kawahara et al. (2018) | 87.1 | 60.0 | 66.1 | 70.9 | 77.2 | 74.2 | 79.7 | 74.2 | 73.7 |
| HcCNN (Bi et al., 2020) | 87.1 | 69.9 | 68.6 | 70.6 | 80.8 | 71.6 | 84.8 | 69.9 | 74.9 |
| FusionM4Net-SS (2022) | 88.1 | 66.1 | 70.1 | 71.1 | 81.5 | 78.0 | 81.8 | 78.5 | 77.0 |
| CC (Wang et al., 2021) | 88.1 | 75.7 | 71.6 | 81.0 | 81.5 | 81.3 | 93.9 | 77.5 | 81.3 |
| Proposed Method (2023) | 91.4 | 76.7 | 78.2 | 81.5 | 81.5 | 81.3 | 94.9 | 78.2 | 83.0 |
Why This Matters: Transforming Skin Cancer Care
This research isn’t just about benchmark scores; it has tangible implications for patient care:
- Enhanced Diagnostic Accuracy: An 83% average accuracy for multi-label classification directly translates to more reliable identification of malignant lesions based on established clinical criteria (the 7-point checklist), leading to earlier and more accurate diagnoses.
- Democratizing Expertise: Such AI systems can act as powerful decision-support tools, particularly in regions with limited access to specialist dermatologists. Mobile implementations could allow for preliminary screening via smartphones.
- Standardizing the SPC: By automating the evaluation of the seven-point checklist criteria, the framework reduces subjectivity and increases consistency in applying this diagnostic standard.
- Efficiency in Clinics: Assisting dermatologists by rapidly pre-screening cases or highlighting critical lesion areas (via attention maps) can streamline workflows and allow experts to focus on complex cases.
If you’re Interested in implementing adversarial defenses in your medical imaging pipeline, you may also find this article helpful: Skin Cancer AI Combats Adversarial Attacks with MDDA
The Future of AI in Dermatology
While this hybrid soft attention framework represents a significant leap forward, research continues. The authors highlight future directions, including:
- Graph Convolutional Networks (GCNs): Integrating GCNs with attention could better model the complex dependencies between different diagnostic labels (e.g., how the presence of “Atypical Pigment Network” influences the likelihood of “Irregular Streaks”).
- Handling Temporal Data: Incorporating sequential dermoscopy images taken over time to track lesion evolution.
- Broader Datasets: Validating and refining the model on larger, more diverse datasets spanning different skin types and ethnicities to improve generalizability.
- Real-World Integration: Developing robust clinical interfaces and workflows to seamlessly integrate such AI tools into dermatologists’ daily practice.
Ethical Considerations: It’s crucial to emphasize that AI is designed to augment dermatologists, not replace them. Final diagnosis and treatment decisions remain the responsibility of qualified medical professionals. Transparency in how AI models arrive at predictions is also essential for building trust.
Conclusion: A Brighter, AI-Assisted Future for Skin Health
The novel hybrid soft attention-based multi-modal deep learning framework developed by researchers at Atatürk University marks a substantial advancement in automated skin lesion diagnosis. By intelligently fusing dermoscopy, clinical images, and patient metadata, and focusing powerfully on relevant lesion features through soft attention, this AI model achieves unprecedented accuracy in multi-label classification against the rigorous seven-point checklist standard.
This breakthrough holds immense promise for improving early skin cancer detection rates globally. As AI continues to evolve, integrating seamlessly with clinical expertise, we move closer to a future where timely and accurate diagnosis is accessible to all, dramatically improving survival rates and patient outcomes.
Call to Action: The Future of Dermatology Starts Here
The proposed soft attention-based AI system for multi-label skin lesion classification is more than just an academic breakthrough—it’s a glimpse into the future of AI-assisted dermatology.
👉 If you’re a healthcare professional, AI researcher, or health-tech innovator, consider exploring or integrating multi-modal AI tools into your workflow.
💬 Interested in trying this model? Connect with the researchers or access the dataset and implementation details on the SPC dataset platform.
📢 Let’s bring precision dermatology to every corner of the world—because early detection saves lives.
Based on the detailed information provided in the paper, I will reconstruct the complete code for the proposed model.
import tensorflow as tf
from tensorflow.keras.layers import (Input, Conv2D, SeparableConv2D, BatchNormalization,
Activation, Add, MaxPooling2D, GlobalAveragePooling2D,
Dense, Concatenate, Multiply, Reshape, Softmax, Lambda)
from tensorflow.keras.models import Model
from tensorflow.keras.activations import relu
import tensorflow.keras.backend as K
def soft_attention_module(input_tensor, k=64):
"""Soft Attention Module (Shaikh et al., 2020) implementation"""
h, w, c = input_tensor.shape[1:]
# Generate attention maps
x = Conv2D(k, (1, 1), padding='same')(input_tensor)
x = Reshape((h*w, k))(x)
x = Softmax(axis=1)(x) # Spatial softmax
x = Reshape((h, w, k))(x)
# Unified attention map
alpha = Lambda(lambda x: K.sum(x, axis=-1, keepdims=True))(x)
# Attention scaling
gamma = tf.Variable(initial_value=0.0, trainable=True)
x_sa = Multiply()([input_tensor, alpha])
x_sa = Lambda(lambda x: x * gamma)(x_sa)
# Residual connection
output = Add()([input_tensor, x_sa])
return output
def entry_flow(input_tensor):
"""Xception entry flow (first 3 blocks)"""
# Block 1
x = Conv2D(32, (3, 3), strides=2, padding='same')(input_tensor)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = Conv2D(64, (3, 3), padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
# Block 2
residual = Conv2D(128, (1, 1), strides=2, padding='same')(x)
residual = BatchNormalization()(residual)
x = SeparableConv2D(128, (3, 3), padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = SeparableConv2D(128, (3, 3), padding='same')(x)
x = BatchNormalization()(x)
x = MaxPooling2D((3, 3), strides=2, padding='same')(x)
x = Add()([x, residual])
# Block 3
residual = Conv2D(256, (1, 1), strides=2, padding='same')(x)
residual = BatchNormalization()(residual)
x = Activation('relu')(x)
x = SeparableConv2D(256, (3, 3), padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = SeparableConv2D(256, (3, 3), padding='same')(x)
x = BatchNormalization()(x)
x = MaxPooling2D((3, 3), strides=2, padding='same')(x)
x = Add()([x, residual])
return x
def middle_flow(input_tensor):
"""Single middle flow block (reduced from 8 to 1)"""
residual = input_tensor
x = Activation('relu')(input_tensor)
x = SeparableConv2D(728, (3, 3), padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = SeparableConv2D(728, (3, 3), padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = SeparableConv2D(728, (3, 3), padding='same')(x)
x = BatchNormalization()(x)
return Add()([x, residual])
def exit_flow(input_tensor):
"""Modified exit flow (without last two separable conv layers)"""
residual = Conv2D(1024, (1, 1), strides=2, padding='same')(input_tensor)
residual = BatchNormalization()(residual)
x = Activation('relu')(input_tensor)
x = SeparableConv2D(728, (3, 3), padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = SeparableConv2D(1024, (3, 3), padding='same')(x)
x = BatchNormalization()(x)
x = MaxPooling2D((3, 3), strides=2, padding='same')(x)
x = Add()([x, residual])
return x
def modified_xception(input_tensor):
"""Modified Xception architecture"""
x = entry_flow(input_tensor) # Output: 19x19x728
x = middle_flow(x) # Output: 19x19x728
x = exit_flow(x) # Output: 10x10x1024
return x
def create_output_heads(x, num_classes):
"""Create 8 output heads for multi-label classification"""
outputs = []
for i, classes in enumerate(num_classes):
outputs.append(Dense(classes, activation='softmax', name=f'output_{i}')(x))
return outputs
def build_model(input_shape=(299, 299, 3), meta_dim=6):
"""Build the complete multi-modal model"""
# Input definitions
dermoscopy_input = Input(shape=input_shape, name='dermoscopy_input')
clinical_input = Input(shape=input_shape, name='clinical_input')
meta_input = Input(shape=(meta_dim,), name='meta_input')
# ====================
# Dermoscopy Branch
# ====================
derm_x = modified_xception(dermoscopy_input)
derm_x = soft_attention_module(derm_x)
derm_x = GlobalAveragePooling2D()(derm_x)
derm_outputs = create_output_heads(derm_x, [3, 2, 3, 3, 3, 3, 2, 5])
# ====================
# Clinical Branch
# ====================
clin_x = modified_xception(clinical_input)
clin_x = soft_attention_module(clin_x)
clin_x = GlobalAveragePooling2D()(clin_x)
clin_outputs = create_output_heads(clin_x, [3, 2, 3, 3, 3, 3, 2, 5])
# ====================
# Hybrid-Meta Branch
# ====================
# Concatenate features from both modalities
derm_features = modified_xception(dermoscopy_input)
clin_features = modified_xception(clinical_input)
concat_features = Concatenate(axis=-1)([derm_features, clin_features])
# Attention path A
attn_a = soft_attention_module(concat_features)
attn_a = GlobalAveragePooling2D()(attn_a)
# Convolutional path B
conv_b = concat_features
for _ in range(2): # Two conv units
conv_b = Conv2D(2048, (3, 3), padding='same')(conv_b)
conv_b = Activation('relu')(conv_b)
conv_b = SeparableConv2D(2048, (3, 3), padding='same')(conv_b)
conv_b = BatchNormalization()(conv_b)
conv_b = soft_attention_module(conv_b)
conv_b = GlobalAveragePooling2D()(conv_b)
# Merge paths and add metadata
merged = Concatenate()([attn_a, conv_b, meta_input])
hybrid_x = Dense(1024, activation='relu')(merged)
hybrid_outputs = create_output_heads(hybrid_x, [3, 2, 3, 3, 3, 3, 2, 5])
# ====================
# Late Fusion
# ====================
fused_outputs = []
for i in range(8):
# Average probabilities from all three branches
avg = tf.keras.layers.Average()([
derm_outputs[i],
clin_outputs[i],
hybrid_outputs[i]
])
fused_outputs.append(avg)
# Create model
model = Model(
inputs=[dermoscopy_input, clinical_input, meta_input],
outputs=fused_outputs,
name='multi_modal_skin_lesion_classifier'
)
return model
# Build and compile the model
model = build_model()
model.summary()
# Compile with appropriate losses for each head
losses = {
'output_0': 'categorical_crossentropy',
'output_1': 'categorical_crossentropy',
'output_2': 'categorical_crossentropy',
'output_3': 'categorical_crossentropy',
'output_4': 'categorical_crossentropy',
'output_5': 'categorical_crossentropy',
'output_6': 'categorical_crossentropy',
'output_7': 'categorical_crossentropy'
}
model.compile(
optimizer=tf.keras.optimizers.Adam(learning_rate=0.0001),
loss=losses,
metrics=['accuracy']
)References
Omeroglu, A. N., Mohammed, H. M. A., Oral, E. A., & Aydin, S. (2023). A novel soft attention-based multi-modal deep learning framework for multi-label skin lesion classification. Engineering Applications of Artificial Intelligence, 120, 105897. https://doi.org/10.1016/j.engappai.2023.105897