Introduction: The Future of Medical Imaging Starts Here
Medical imaging has long been a cornerstone of diagnostics, but traditional methods often fall short when it comes to adapting to real-world variability. Enter counterfactual contrastive learning , an innovative framework that’s changing the game by leveraging causal image synthesis to improve model robustness and downstream performance.
In this article, we’ll explore how counterfactual contrastive learning outperforms standard contrastive learning, especially under limited labels and for images acquired with scanners under-represented in the training set. We’ll also show you why adopting this method could be one of the most impactful decisions for your practice.
What Is Counterfactual Contrastive Learning?
A New Paradigm in Self-Supervised Learning
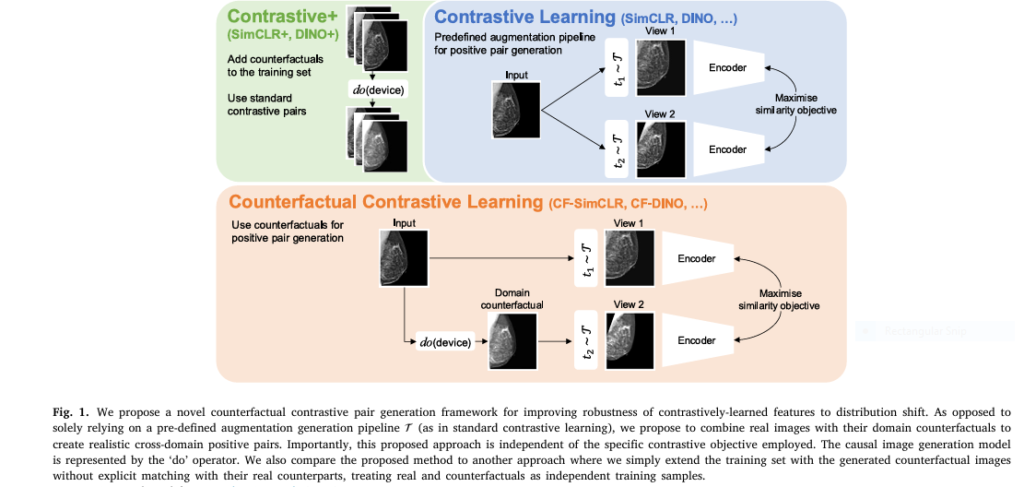
Contrastive pretraining is already known for boosting generalization and downstream performance in machine learning models. However, its effectiveness hinges on the data augmentation strategy used to generate positive pairs. Most traditional approaches rely on generic image transformations like color jittering or rotation—techniques borrowed from natural image processing.
But here’s the problem: these methods don’t always mimic realistic domain variations found in medical imaging, such as differences between MRI scanners or X-ray machines.
That’s where counterfactual contrastive learning steps in. This novel approach uses causal image synthesis to create contrastive positive pairs that better reflect real-world domain shifts. By simulating scanner changes or other acquisition differences, the model learns more robust representations that generalize across devices and settings.
How Does It Work? A Technical Breakdown
Step-by-Step Process Behind the Innovation
- Causal Image Generation Model : At the heart of counterfactual contrastive learning lies a deep structural causal model (DSCM) that generates high-quality, realistic counterfactual images. These are synthetic images that answer “what-if” questions—like what a mammogram would look like if it were captured using a different scanner.
- Positive Pair Creation : Instead of applying random augmentations, the system matches real images with their domain-specific counterfactuals. For example, an X-ray taken on Scanner A is paired with a synthesized version of the same image as if it had been captured on Scanner B.
- Contrastive Objective : The model then learns to bring similar representations closer while pushing dissimilar ones apart, just like in SimCLR or DINO-v2 frameworks—but now with more meaningful and realistic variation.
- Robust Feature Learning : As a result, the learned features are less sensitive to acquisition-related noise and more focused on clinically relevant patterns.
Why It Outperforms Standard Methods
Key Advantages Over Traditional Contrastive Learning
✅ Superior Generalization Across Domains
Traditional contrastive learning struggles when test data differs significantly from training data—especially in terms of scanner types or imaging protocols. Counterfactual contrastive learning addresses this by explicitly modeling domain shifts during training.
Example : On the PadChest dataset, models trained with counterfactual contrastive learning showed improved performance on external datasets like RSNA Pneumonia and CheXpert, even though those domains weren’t seen during pretraining.
✅ Better Performance on Underrepresented Scanners
In many clinical environments, certain scanners are used far less frequently than others. Standard models tend to perform poorly on these underrepresented domains due to lack of exposure.
With counterfactual contrastive learning, synthetic domain variations ensure balanced training across all scanner types—even those with very few real samples.
Result : In experiments, underrepresented scanners saw up to a 6% improvement in ROC-AUC compared to baseline models.
✅ Reduces Subgroup Disparities (e.g., Biological Sex)
Beyond scanner differences, counterfactual contrastive learning can also be applied to reduce disparities across biological sex, age groups, or ethnicities. By generating cross-subgroup pairs (e.g., male-to-female counterfactuals), the model learns fairer, more balanced representations.
Impact : On pneumonia detection tasks, female subgroups saw up to a 5% improvement in diagnostic accuracy , closing the gap with male patients.
✅ Aggressive Data Efficiency
One of the biggest challenges in medical AI is the scarcity of labeled data. Counterfactual contrastive learning shines in low-data regimes.
Evidence : On EMBED mammography data, models trained with only 2,230 labeled samples achieved nearly the same performance as fully supervised models trained on over 200,000 images .
✅ Flexible & Compatible with Modern Architectures
Whether you’re using SimCLR , DINO-v2 , or any modern vision transformer architecture, counterfactual contrastive learning integrates seamlessly. It doesn’t replace your existing contrastive objective—it enhances it.
Real-World Applications: Where It Makes the Biggest Impact
🏥 Radiology: Improving Chest X-Ray Diagnostics
Chest radiography is one of the most widely used diagnostic tools globally. However, variations in equipment, technique, and post-processing can severely impact model performance.
By training models with counterfactual contrastive learning:
- Accuracy improves across multiple external datasets
- Models become more robust to scanner differences
- Diagnostic consistency increases across hospitals and clinics
🩺 Mammography: Enhancing Breast Cancer Detection
Breast density assessment and cancer detection are highly dependent on image quality and scanner type. Counterfactual contrastive learning ensures that models trained on dominant scanner types (like Selenia Dimensions) still perform well on rarer ones (like Senographe Pristina).
Outcome : Up to 4% increase in sensitivity on underrepresented scanners, improving early detection rates.
🧠 Neuroimaging: Addressing Bias in Brain Scan Analysis
Bias in brain imaging models can lead to misdiagnosis in underrepresented populations. By generating counterfactuals based on demographic variables (e.g., race, gender), models become more equitable and reliable.
Case Study: Comparative Performance on Five Medical Datasets
| DATASET | METHOD | METRIC IMPROVEMENT |
|---|---|---|
| PadChest | CF-SimCLR vs. SimCLR | +2.5% on Imaging, +0.6% on Phillips |
| CheXpert | CF-SimCLR vs. SimCLR | +1–2% ROC-AUC |
| EMBED | CF-SimCLR vs. SimCLR | +3% on Clearview CSM |
| VinDR Mammo | CF-SimCLR vs. SimCLR | +4–6% ROC-AUC |
| RSNA Pneumonia | CF-DINO vs. DINO | +3.5% with 2158 labels |
These results highlight consistent improvements across both linear probing and full model finetuning , proving that counterfactual contrastive learning isn’t just a theoretical advantage—it delivers real-world value.
If you’re Interested in advance methods in Breast Cancer Radiotherapy with Large language Model, you may also find this article helpful: Beyond Human Limits 1: How RO-LMM’s AI is Revolutionizing Breast Cancer Radiotherapy Planning (Saving Lives & Time)
FAQ: Answering the Questions You’re Searching For
❓ What is contrastive learning in medical imaging?
Contrastive learning is a self-supervised method that trains models by comparing similar (positive) and dissimilar (negative) image pairs. In medical imaging, this helps models learn meaningful representations without requiring large amounts of labeled data.
❓ How does counterfactual contrastive learning differ from standard contrastive learning?
While standard contrastive learning uses generic image transformations (e.g., rotations, crops), counterfactual contrastive learning uses realistic domain shifts (e.g., scanner changes) generated via causal image synthesis. This leads to more robust and generalizable models.
❓ Why is counterfactual contrastive learning important for healthcare AI?
It improves model fairness, reduces scanner dependency, enhances performance on underrepresented populations, and requires fewer labeled images—making it ideal for real-world deployment.
❓ Which medical imaging modalities benefit most from this technique?
Chest radiography and mammography have shown the most significant improvements, but the method is applicable to MRI, CT, and ultrasound as well.
Call to Action: Ready to Transform Your Medical Imaging Workflow?
If you’re working in radiology, pathology, or any field that relies on medical imaging, now is the time to explore counterfactual contrastive learning . Whether you’re developing internal AI tools or evaluating vendor solutions, understanding this technology can give you a competitive edge and improve patient outcomes.
👉 Download the full research paper here to dive deeper into the technical details and implementation strategies.
👉 For complete training scripts and pretrained models, visit the official repository:
biomedia-mira/counterfactual-contrastive
👉 Schedule a consultation with our team of medical AI experts to see how this approach can be integrated into your current pipeline.
Final Thoughts: Embracing Innovation in Medical AI
The future of medical imaging is not just about higher resolution or faster scans—it’s about smarter models that adapt to real-world variability . Counterfactual contrastive learning offers a path forward by combining the best of generative modeling and contrastive learning to build AI systems that are robust, fair, and efficient .
As the demand for accurate, scalable, and ethical AI grows, so too will the importance of techniques like counterfactual contrastive learning. Don’t get left behind—start exploring how this powerful method can revolutionize your imaging workflows today.
Complete Implementation of Counterfactual Contrastive Learning.
import torch
import torch.nn as nn
import torch.nn.functional as F
from torchvision import transforms
from torch.utils.data import Dataset, DataLoader
from einops import rearrange
import numpy as np
# ======================
# 1. Hierarchical VAE (HVAE) for Counterfactual Generation
# ======================
class ResidualBlock(nn.Module):
"""Basic residual block with GroupNorm and SiLU activation"""
def __init__(self, in_channels, out_channels):
super().__init__()
self.conv1 = nn.Conv2d(in_channels, out_channels, 3, padding=1)
self.conv2 = nn.Conv2d(out_channels, out_channels, 3, padding=1)
self.group_norm1 = nn.GroupNorm(8, in_channels)
self.group_norm2 = nn.GroupNorm(8, out_channels)
self.silu = nn.SiLU()
self.residual = nn.Conv2d(in_channels, out_channels, 1) if in_channels != out_channels else nn.Identity()
def forward(self, x):
residual = self.residual(x)
x = self.group_norm1(x)
x = self.silu(x)
x = self.conv1(x)
x = self.group_norm2(x)
x = self.silu(x)
x = self.conv2(x)
return x + residual
class HVAE_Encoder(nn.Module):
"""Hierarchical VAE Encoder with multiple stochastic layers"""
def __init__(self, z_dims=[64, 32, 16], input_channels=1):
super().__init__()
self.blocks = nn.ModuleList()
self.z_dims = z_dims
in_ch = input_channels
# Downsampling path
for i, dim in enumerate(z_dims):
# Residual blocks
self.blocks.append(nn.Sequential(
ResidualBlock(in_ch, dim),
ResidualBlock(dim, dim)
))
# Downsample
if i < len(z_dims) - 1:
self.blocks.append(nn.Conv2d(dim, dim, 3, stride=2, padding=1))
in_ch = dim
# Final layers for mean and logvar
self.fc_mu = nn.ModuleList()
self.fc_logvar = nn.ModuleList()
for dim in z_dims:
self.fc_mu.append(nn.Conv2d(dim, dim, 1))
self.fc_logvar.append(nn.Conv2d(dim, dim, 1))
def forward(self, x, parents):
# Parent conditioning (e.g., scanner type or sex)
parent_embed = parents.unsqueeze(-1).unsqueeze(-1).expand(-1, -1, *x.shape[2:])
x = torch.cat([x, parent_embed], dim=1)
# Hierarchical encoding
mus, logvars = [], []
for i, block in enumerate(self.blocks):
x = block(x)
if isinstance(block, nn.Sequential): # After residual blocks
mu = self.fc_mu[i//2](x)
logvar = self.fc_logvar[i//2](x)
mus.append(mu)
logvars.append(logvar)
return mus, logvars
class HVAE_Decoder(nn.Module):
"""Hierarchical VAE Decoder with skip connections"""
def __init__(self, z_dims=[16, 32, 64], output_channels=1):
super().__init__()
self.z_dims = z_dims[::-1]
self.blocks = nn.ModuleList()
# Initial projection
self.init_conv = nn.Conv2d(self.z_dims[0], self.z_dims[0], 1)
# Upsampling path
for i, dim in enumerate(self.z_dims):
self.blocks.append(ResidualBlock(dim, dim))
if i < len(self.z_dims) - 1:
self.blocks.append(nn.ConvTranspose2d(dim, self.z_dims[i+1], 4, stride=2, padding=1))
# Final output
self.final_conv = nn.Conv2d(self.z_dims[-1], output_channels, 3, padding=1)
self.parent_fc = nn.Linear(1, self.z_dims[0]) # Parent conditioning
def forward(self, z_list, parents):
# Parent conditioning
parent_embed = self.parent_fc(parents).unsqueeze(-1).unsqueeze(-1)
x = z_list[0] + parent_embed
for block in self.blocks:
x = block(x)
# Add skip connections
if isinstance(block, ResidualBlock) and block == self.blocks[0]:
for z in z_list[1:]:
x = x + F.interpolate(z, size=x.shape[2:], mode='nearest')
x = self.final_conv(x)
return torch.sigmoid(x)
class HVAE(nn.Module):
"""Full Hierarchical Variational Autoencoder"""
def __init__(self, z_dims=[64, 32, 16], input_channels=1):
super().__init__()
self.encoder = HVAE_Encoder(z_dims, input_channels)
self.decoder = HVAE_Decoder(z_dims, input_channels)
self.z_dims = z_dims
def reparameterize(self, mu, logvar):
std = torch.exp(0.5*logvar)
eps = torch.randn_like(std)
return mu + eps*std
def forward(self, x, parents):
# Encode
mus, logvars = self.encoder(x, parents)
# Reparameterize
z_list = [self.reparameterize(mu, logvar) for mu, logvar in zip(mus, logvars)]
# Decode
recon = self.decoder(z_list, parents)
return recon, mus, logvars
def generate_counterfactual(self, x, orig_parents, target_parents):
"""Generate counterfactual image by changing parent variable"""
with torch.no_grad():
mus, logvars = self.encoder(x, orig_parents)
z_list = [self.reparameterize(mu, logvar) for mu, logvar in zip(mus, logvars)]
return self.decoder(z_list, target_parents)
# ======================
# 2. Counterfactual Contrastive Learning
# ======================
class ProjectionHead(nn.Module):
"""Projection head for contrastive learning"""
def __init__(self, input_dim=512, hidden_dim=256, output_dim=128):
super().__init__()
self.layers = nn.Sequential(
nn.Linear(input_dim, hidden_dim),
nn.BatchNorm1d(hidden_dim),
nn.ReLU(),
nn.Linear(hidden_dim, output_dim)
def forward(self, x):
return F.normalize(self.layers(x), dim=-1)
class CF_SimCLR(nn.Module):
"""Counterfactual Contrastive Learning with SimCLR Framework"""
def __init__(self, encoder, h_vae, parent_dim=1):
super().__init__()
self.encoder = encoder # ResNet or ViT backbone
self.projection = ProjectionHead()
self.h_vae = h_vae
self.parent_dim = parent_dim
self.augment = transforms.Compose([
transforms.RandomHorizontalFlip(),
transforms.RandomResizedCrop(size=224),
transforms.ColorJitter(0.2, 0.2, 0.2, 0.1),
transforms.GaussianBlur(3)
])
def forward(self, x, parents):
# Generate counterfactuals
target_parents = torch.randint(0, self.parent_dim, (x.size(0), device=x.device)
cf_x = self.h_vae.generate_counterfactual(x, parents, target_parents)
# Create positive pairs
x1 = self.augment(x)
x2 = self.augment(cf_x)
# Extract features
z1 = self.projection(self.encoder(x1))
z2 = self.projection(self.encoder(x2))
return z1, z2
def nt_xent_loss(self, z1, z2, temperature=0.1):
"""Normalized Temperature-scaled Cross Entropy Loss"""
batch_size = z1.size(0)
z = torch.cat([z1, z2], dim=0)
# Similarity matrix
sim = torch.mm(z, z.t()) / temperature
sim.fill_diagonal_(-float('inf'))
# Positive pairs
pos_mask = torch.eye(batch_size, dtype=torch.bool, device=z.device)
pos_mask = pos_mask.repeat(2, 2)
pos_mask.fill_diagonal_(False)
# Compute loss
pos = sim[pos_mask].reshape(2*batch_size, 1)
neg = sim[~pos_mask].reshape(2*batch_size, -1)
logits = torch.cat([pos, neg], dim=1)
labels = torch.zeros(2*batch_size, dtype=torch.long, device=z.device)
return F.cross_entropy(logits, labels)
# ======================
# 3. DINO-v2 Integration
# ======================
class CF_DINO(nn.Module):
"""Counterfactual Contrastive Learning with DINO-v2 Framework"""
def __init__(self, encoder, h_vae, parent_dim=1):
super().__init__()
self.student = encoder
self.teacher = copy.deepcopy(encoder)
self.h_vae = h_vae
self.parent_dim = parent_dim
# Freeze teacher
for param in self.teacher.parameters():
param.requires_grad = False
self.student_proj = nn.Linear(encoder.embed_dim, 256)
self.teacher_proj = nn.Linear(encoder.embed_dim, 256)
# Global and local views
self.global_crop = transforms.Compose([
transforms.RandomResizedCrop(224, scale=(0.8, 1.0)),
transforms.RandomHorizontalFlip()
])
self.local_crop = transforms.Compose([
transforms.RandomResizedCrop(96, scale=(0.3, 0.8)),
transforms.RandomHorizontalFlip()
])
def generate_views(self, x, parents):
"""Generate global and local views with counterfactuals"""
target_parents = torch.randint(0, self.parent_dim, (x.size(0)), device=x.device)
cf_x = self.h_vae.generate_counterfactual(x, parents, target_parents)
# Global views
global1 = self.global_crop(x)
global2 = self.global_crop(cf_x)
# Local views
local_views = [self.local_crop(x) for _ in range(4)] + \
[self.local_crop(cf_x) for _ in range(4)]
return global1, global2, local_views
def forward(self, x, parents):
# Generate views
global1, global2, local_views = self.generate_views(x, parents)
all_views = [global1, global2] + local_views
# Student embeddings
student_out = [self.student_proj(self.student(view)) for view in all_views]
student_out = [F.normalize(out, dim=-1) for out in student_out]
# Teacher embeddings (only global views)
with torch.no_grad():
teacher_out1 = F.normalize(self.teacher_proj(self.teacher(global1)), dim=-1)
teacher_out2 = F.normalize(self.teacher_proj(self.teacher(global2)), dim=-1)
teacher_out = torch.cat([teacher_out1, teacher_out2])
return student_out, teacher_out
def dino_loss(self, student_out, teacher_out, center_momentum=0.9):
"""DINO-v2 loss with centering"""
# Center teacher outputs
teacher_center = teacher_out.mean(dim=0)
teacher_out = teacher_out - teacher_center
# Compute loss
loss = 0
for s in student_out:
loss += -torch.mean(torch.sum(s * teacher_out, dim=-1))
# Update center
self.center = self.center * center_momentum + teacher_center * (1 - center_momentum)
return loss / len(student_out)
# ======================
# 4. Data Pipeline
# ======================
class MedicalDataset(Dataset):
def __init__(self, image_paths, parent_labels, transform=None):
self.image_paths = image_paths
self.parent_labels = parent_labels
self.transform = transform or transforms.Compose([
transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor()
])
def __len__(self):
return len(self.image_paths)
def __getitem__(self, idx):
img = Image.open(self.image_paths[idx]).convert('L') # Grayscale
parent = self.parent_labels[idx]
return self.transform(img), torch.tensor(parent, dtype=torch.float)
# ======================
# 5. Training Loop
# ======================
def train_hvae(model, dataloader, epochs=50, device='cuda'):
"""Train Hierarchical VAE for counterfactual generation"""
model.to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=1e-4)
for epoch in range(epochs):
for images, parents in dataloader:
images, parents = images.to(device), parents.to(device)
# Forward pass
recon, mus, logvars = model(images, parents)
# Loss components
recon_loss = F.mse_loss(recon, images, reduction='sum')
kl_loss = sum([-0.5 * torch.sum(1 + logvar - mu.pow(2) - logvar.exp())
for mu, logvar in zip(mus, logvars)])
loss = recon_loss + 0.001 * kl_loss
# Backpropagation
optimizer.zero_grad()
loss.backward()
optimizer.step()
print(f'Epoch {epoch+1}/{epochs} | Loss: {loss.item():.4f}')
def train_ccl(model, dataloader, epochs=100, device='cuda'):
"""Train counterfactual contrastive model"""
model.to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=3e-4)
for epoch in range(epochs):
for images, parents in dataloader:
images, parents = images.to(device), parents.to(device)
if isinstance(model, CF_SimCLR):
# SimCLR-style training
z1, z2 = model(images, parents)
loss = model.nt_xent_loss(z1, z2)
elif isinstance(model, CF_DINO):
# DINO-style training
student_out, teacher_out = model(images, parents)
loss = model.dino_loss(student_out, teacher_out)
# Optimization
optimizer.zero_grad()
loss.backward()
optimizer.step()
# Update teacher weights (for DINO)
if isinstance(model, CF_DINO):
with torch.no_grad():
for s_param, t_param in zip(model.student.parameters(),
model.teacher.parameters()):
t_param.data.mul_(0.996).add_(s_param.data * (1 - 0.996))
print(f'Epoch {epoch+1}/{epochs} | Loss: {loss.item():.4f}')
# ======================
# 6. Usage Example
# ======================
if __name__ == "__main__":
# Initialize models
hvae = HVAE(z_dims=[64, 32, 16], input_channels=1)
encoder = torch.hub.load('facebookresearch/dino:main', 'dino_vits16') # ViT-Small
# Prepare dataset (example)
dataset = MedicalDataset(image_paths, scanner_labels) # Implement your own
dataloader = DataLoader(dataset, batch_size=64, shuffle=True)
# 1. Train HVAE first
train_hvae(hvae, dataloader, epochs=50)
# 2. Train CF-SimCLR
cf_simclr = CF_SimCLR(encoder, hvae, parent_dim=N_SCANNERS)
train_ccl(cf_simclr, dataloader, epochs=100)
# Or train CF-DINO
cf_dino = CF_DINO(encoder, hvae, parent_dim=N_SCANNERS)
train_ccl(cf_dino, dataloader, epochs=100)References
Roschewitz M. et al. (2025). Robust Image Representations with Counterfactual Contrastive Learning. Medical Image Analysis 105, 103668.
Azizi S. et al. (2023). Robust and data-efficient generalization… Nature Biomedical Engineering.
Ribeiro F.D.S. et al. (2023). High fidelity image counterfactuals… ICML.

Pingback: Revolutionary AI Breakthrough: Non-Contrast Tumor Segmentation Saves Lives & Avoids Deadly Risks - aitrendblend.com