Imagine finding a single unique galaxy among 100 million images—a cosmic needle in a haystack. This daunting task faces astronomers daily. But what if an AI could pinpoint these rarities while slashing human review time by 90%? Enter AnomalyMatch, the breakthrough framework transforming anomaly detection in astronomy, medical imaging, industrial inspection, and beyond.
The Anomaly Detection Crisis
Identifying statistically unusual objects—like defective products on a factory line, rare diseases in medical scans, or novel celestial phenomena—is critical across industries. Yet traditional methods hit major roadblocks:
- Supervised learning demands thousands of labeled anomalies—often impossible to obtain.
- Unsupervised methods (like Isolation Forests) struggle with complex image data, drowning in false positives.
- Human review of massive datasets is prohibitively slow and costly.
“In astronomy alone, upcoming surveys like Euclid will generate billions of images. Finding scientifically valuable anomalies manually is like drinking from a firehose,” explains Pablo Gomez, ESA scientist and co-creator of AnomalyMatch.
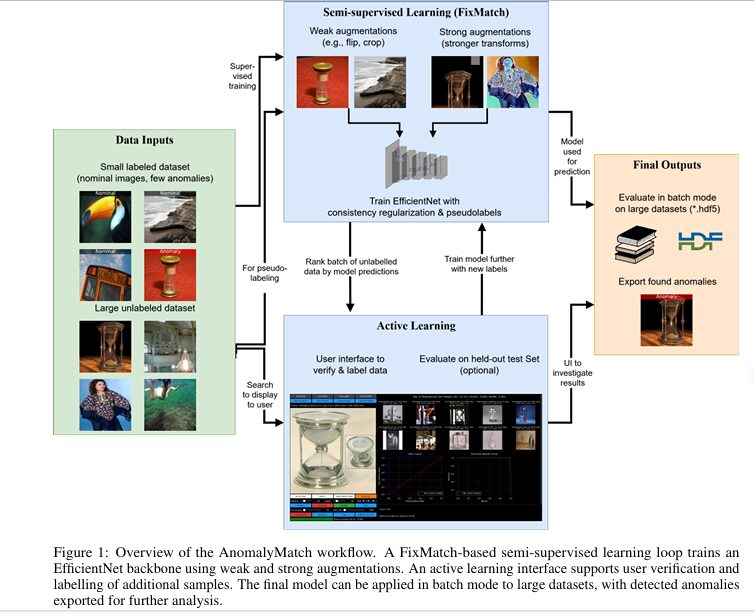
Introducing AnomalyMatch: The Hybrid Solution
Developed by the European Space Agency, AnomalyMatch fuses three powerful techniques into a single scalable workflow:
- Semi-Supervised Learning (FixMatch Algorithm): Uses minimal labeled data + abundant unlabeled images
- Active Learning: Iteratively queries human experts to label high-value uncertainties
- EfficientNet Backbone: Processes images 5x faster than ResNet alternatives
This trifecta tackles the core challenges of rare object discovery: extreme class imbalance (e.g., 1 anomaly per 10,000 images), label scarcity, and computational burden.
How It Works: The Technical Breakthrough
- Binary Classification Setup:
- Trains a model to distinguish “normal” vs. “anomaly” classes
- Starts with just 5–10 labeled anomalies (e.g., unusual galaxy types)
- FixMatch Optimization:
- Generates “pseudo-labels” for unlabelled data via weak/strong augmentations
- Enforces prediction consistency: If weak-augmented and strong-augmented versions of an image agree (confidence >95%), it becomes training data
- Active Learning GUI:
- Ranks images by anomaly score
- Experts label top candidates + correct false positives
- Model retrains in hours, improving precision iteratively
“Unlike black-box AI, our interface lets astronomers steer the search toward scientifically interesting anomalies—not just statistical outliers,” notes co-developer David O’Ryan.
Performance That Redefines Expectations
Tested on GalaxyMNIST (astronomy) and miniImageNet (general images) with severe class imbalance:
| Metric | miniImageNet (1% anomalies) | GalaxyMNIST (25% anomalies) |
|---|---|---|
| AUROC | 0.95 | 0.86 |
| AUPRC | 0.77 | 0.71 |
| Anomalies in Top 1% | 71–81% | Up to 98% (Unbarred Spirals) |
| Processing Speed | 100M images in 3 days (1 GPU) |
Real-World Impact:
- After 3 active learning cycles (adding just 10 labels/cycle), 93% precision for top-ranked anomalies in GalaxyMNIST
- Reduces manual review volume by 99.9%—experts inspect only the top 0.1% of predictions
5 Industries Revolutionized by AnomalyMatch
- Astronomy:
- Scan Hubble Legacy Archive images for rare galaxies, asteroids, or artifacts
- Integrated into ESA’s Datalabs for Euclid mission analysis
- Medical Imaging:
- Detect rare tumors in MRI/CT scans with minimal labeled data
- Manufacturing:
- Identify micro-defects in semiconductor wafers or automotive parts
- Environmental Monitoring:
- Spot illegal deforestation or pollution sources in satellite imagery
- Security:
- Recognize suspicious objects in airport scans with fewer false alarms
If you’re Interested in purely FixMatch implementation, you may also find this article helpful: FixMatch: Simplified SSL Breakthrough
Limitations and Future Frontiers
While groundbreaking, challenges remain:
- Labeling errors during active learning may propagate bias
- Extreme imbalance (<0.01% anomalies) needs further testing
- Heterogeneous anomalies (multiple anomaly types) require architecture tweaks
Next-Gen Upgrades in Development:
- Explainable AI: SHAP values to clarify why an object is flagged
- Multimodal Analysis: Combine images with spectral/temporal data
- Vision Transformers: Boosting accuracy on texture-rich anomalies
- Noise-Robust Training: Mitigating human labeling errors
The Future of Discovery Starts Now
AnomalyMatch proves that scarce labels and massive datasets no longer bottleneck discovery. By merging human intuition with AI scalability, it turns the impossible into the actionable.
For Astronomers & Data Scientists:
The framework will be open-sourced on ESA’s GitHub pending licensing. Preprint available on arXiv.
For Enterprises:
Custom deployments can process proprietary datasets—from detecting financial fraud to discovering new materials.
“We’re entering an era where finding one-in-a-million objects takes days, not lifetimes,” says Gomez. “This is just the start.”
Ready to Transform Your Anomaly Detection?
➔ Astronomy Teams: Request early access via ESA’s Datalabs platform
➔ Industry Leaders: Contact ESA’s Tech Transfer Office for licensing
➔ Developers: Star the GitHub repo to get release updates
Unlock the rare. Scale the impossible.
Below is a code implementation of the AnomalyMatch model. This code is a simplified version and may need to be adjusted based on specific requirements and environments.
import torch
import torch.nn as nn
import torch.optim as optim
from torchvision import models, transforms
from torch.utils.data import Dataset, DataLoader
import numpy as np
from PIL import Image
import os
import glob
# Define the AnomalyMatch model
class AnomalyMatch(nn.Module):
def __init__(self, num_classes=2):
super(AnomalyMatch, self).__init__()
self.efficientnet = models.efficientnet_b0(pretrained=True)
self.efficientnet.classifier = nn.Linear(self.efficientnet.classifier[1].in_features, num_classes)
def forward(self, x):
return self.efficientnet(x)
# Define the dataset
class AnomalyDataset(Dataset):
def __init__(self, image_paths, labels=None, transform=None):
self.image_paths = image_paths
self.labels = labels
self.transform = transform
def __len__(self):
return len(self.image_paths)
def __getitem__(self, idx):
image = Image.open(self.image_paths[idx])
if self.transform:
image = self.transform(image)
if self.labels is not None:
label = self.labels[idx]
return image, label
else:
return image, idx
# Define data augmentation
def get_data_transforms():
weak_transforms = transforms.Compose([
transforms.Resize((224, 224)),
transforms.RandomHorizontalFlip(),
transforms.RandomCrop(224, padding=4),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
strong_transforms = transforms.Compose([
transforms.Resize((224, 224)),
transforms.RandomHorizontalFlip(),
transforms.RandomCrop(224, padding=4),
transforms.ColorJitter(brightness=0.5, contrast=0.5, saturation=0.5, hue=0.1),
transforms.RandomRotation(20),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
eval_transforms = transforms.Compose([
transforms.Resize((224, 224)),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
return weak_transforms, strong_transforms, eval_transforms
# Training function
def train(model, dataloader, criterion, optimizer, device, confidence_threshold=0.95):
model.train()
total_loss = 0
for images, labels in dataloader:
images, labels = images.to(device), labels.to(device)
weak_augmented_images = images
strong_augmented_images = images
optimizer.zero_grad()
weak_outputs = model(weak_augmented_images)
strong_outputs = model(strong_augmented_images)
supervised_loss = criterion(weak_outputs, labels)
pseudo_labels = torch.argmax(strong_outputs.detach(), dim=1)
pseudo_label_mask = torch.max(strong_outputs.detach(), dim=1)[0] >= confidence_threshold
unsupervised_loss = criterion(strong_outputs[pseudo_label_mask], pseudo_labels[pseudo_label_mask])
loss = supervised_loss + unsupervised_loss
loss.backward()
optimizer.step()
total_loss += loss.item()
return total_loss / len(dataloader)
# Active learning loop
def active_learning_loop(model, unlabeled_dataset, device, num_samples=10):
model.eval()
anomaly_scores = []
with torch.no_grad():
for images, idx in unlabeled_dataset:
images = images.to(device)
outputs = model(images)
scores = torch.softmax(outputs, dim=1)[:, 1].cpu().numpy()
anomaly_scores.extend(scores)
anomaly_scores = np.array(anomaly_scores)
top_indices = np.argsort(anomaly_scores)[-num_samples:]
return top_indices
# Main function
if __name__ == "__main__":
# Hyperparameters
batch_size = 16
learning_rate = 0.0075
num_epochs = 10
confidence_threshold = 0.95
# Device configuration
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# Create the model
model = AnomalyMatch(num_classes=2)
model = model.to(device)
# Define loss function and optimizer
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(model.parameters(), lr=learning_rate, momentum=0.9, weight_decay=7.5e-4)
# Load datasets
labeled_image_paths = glob.glob("labeled_data/*")
labeled_labels = [0] * len(labeled_image_paths) # Replace with actual labels
unlabeled_image_paths = glob.glob("unlabeled_data/*")
weak_transforms, strong_transforms, eval_transforms = get_data_transforms()
labeled_dataset = AnomalyDataset(labeled_image_paths, labeled_labels, transform=weak_transforms)
labeled_dataloader = DataLoader(labeled_dataset, batch_size=batch_size, shuffle=True)
unlabeled_dataset = AnomalyDataset(unlabeled_image_paths, transform=strong_transforms)
unlabeled_dataloader = DataLoader(unlabeled_dataset, batch_size=batch_size, shuffle=True)
# Training loop
for epoch in range(num_epochs):
train_loss = train(model, labeled_dataloader, criterion, optimizer, device, confidence_threshold)
print(f"Epoch {epoch+1}, Loss: {train_loss:.4f}")
# Active learning
if epoch % 3 == 0:
top_indices = active_learning_loop(model, unlabeled_dataloader, device, num_samples=10)
for idx in top_indices:
# Label the selected samples and add them to the labeled dataset
# This part requires human interaction to label the samples
passIf you want access the complete paper, then click this link: https://arxiv.org/abs/2504.03705