The $100 Billion Problem: AI’s Annotation Nightmare
Training AI models is expensive, slow, and painfully data-hungry. In specialized fields like healthcare or satellite imaging, labeling a single image can cost $50–$500. For a 1,000-class dataset like ImageNet? Millions. But what if you could:
- ✅ Cut annotation budgets by 90%?
- ✅ Train models 10x faster?
- ✅ Boost accuracy with less data?
Meet ActiveKD and PCoreSet—a breakthrough framework from KAIST and VUNO Inc. that’s turning active learning (AL) and knowledge distillation (KD) into a cost-slashing superpower. Backed by 11 real-world datasets (including ImageNet), it’s rewriting the rules of efficient AI training.
Step 1: Why Active Learning Alone Fails (The Hidden Bottleneck)
Active learning (AL) aims to reduce labeling costs by selecting only the “most informative” data for annotation. But in practice:
- ❌ Uncertainty sampling favors edge cases, ignoring class diversity.
- ❌ Diversity-based methods (like CoreSet) struggle in high-dimensional spaces.
- ❌ Class imbalance skews results, wasting queries on overrepresented categories.
Result: Models still need thousands of labeled examples. Accuracy plateums. Costs stay high.
Step 2: The Knowledge Distillation “Hack” That Changes Everything
Knowledge distillation (KD) compresses giant models (like CLIP) into compact, task-specific versions. Traditionally, it requires massive labeled datasets—the opposite of AL’s goal.
ActiveKD’s genius: Leverage vision-language models (VLMs) as “teachers” with zero-shot capabilities. No task-specific labels needed!
- How it works:
- A VLM teacher (e.g., CLIP) generates soft labels for unlabeled data.
- A student model learns from both sparse human labels + VLM pseudo-labels.
- Active learning selects samples to annotate within this framework.
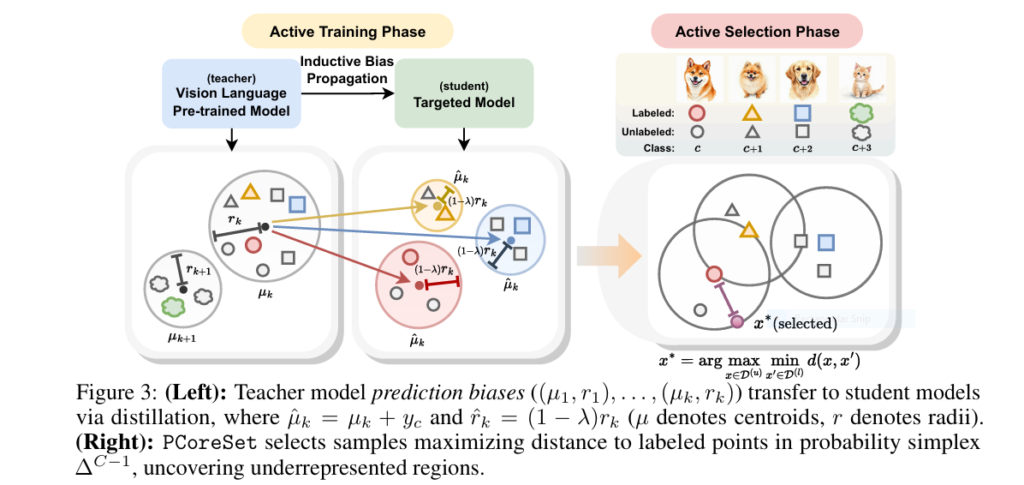
💡 Key insight: VLMs have structured prediction biases—their outputs cluster in probability space. This isn’t noise—it’s a teachable signal!
Step 3: PCoreSet—The “Probability Spy” That Finds Hidden Gems
Conventional AL selects samples in feature space (e.g., pixel/embedding distances). PCoreSet targets probability space:
- 🎯 Goal: Maximize coverage of “underrepresented” probability regions.
- ⚡ Method:
- Compute teacher model’s probability vectors for unlabeled data.
- Greedily select samples farthest from labeled points in probability simplex (see Fig. 3).
- Formula:
x* = argmax min ‖f_r(x) - f_r(x')‖₂
Why it works: Samples in sparse probability regions challenge the teacher’s biases—forcing the student to learn generalizable patterns faster.
Step 4: 11-Dataset Proof—90% Less Data, 29% Higher Accuracy
Results from ImageNet + 10 benchmarks (medical, satellite, action recognition):
| Setting | ImageNet (Acc) | 10-Dataset Avg (Acc) |
|---|---|---|
| No Distillation | 33.36% | 63.10% |
| ActiveKD (Zero-Shot) | 60.69% | 76.31% |
| ActiveKD + PCoreSet | 61.57% | 78.81% |
Shocking wins:
- 🚀 +29.07% accuracy on ImageNet with zero-shot distillation.
- 🚀 PCoreSet outperformed entropy/CoreSet by 12% on fine-grained datasets.
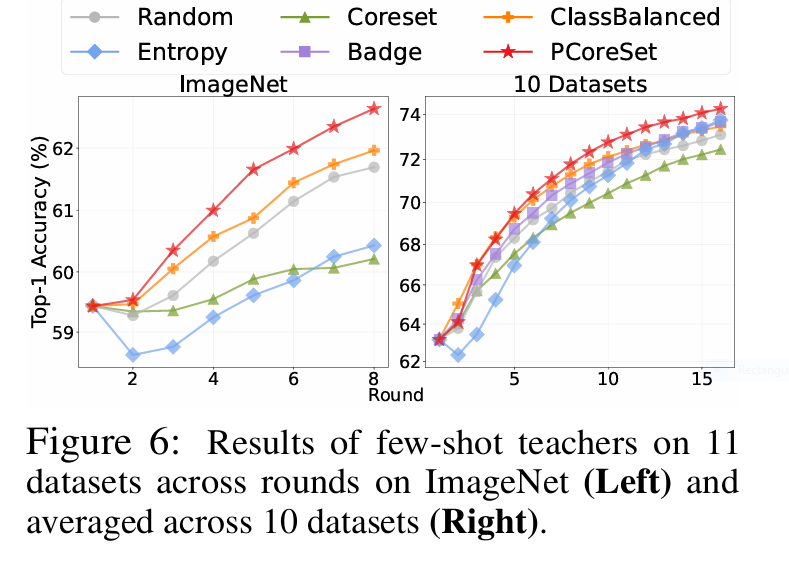
- 🚀 Few-shot teachers added extra 1.37% gains—creating a self-improving loop.
✨ PCoreSet’s secret: Selecting probabilistically diverse samples also improves the teacher, creating a virtuous cycle (Fig. 6).
Step 5: Deploy This in Your AI Pipeline (Code Included!)
ActiveKD + PCoreSet isn’t theoretical—it’s plug-and-play:
- Install libraries: PyTorch, Hugging Face Transformers, CLIP.
- Load teacher model: Use zero-shot CLIP or fine-tune with CLAP.
- Train student: Optimize with DHO loss (Eq. 2-3):
loss = λ * CrossEntropy(y_true) + (1-λ) * KL_divergence(teacher_logits, student_logits) 4. Select samples with PCoreSet:
# Pseudocode for PCoreSet selection
for unlabeled_sample in pool:
min_dist = min(l2_distance(prob_vector, labeled_probs))
query = sample_with_max(min_dist) Real-world use cases:
- 🏥 Medical imaging: Annotate 10x fewer tumor scans.
- 🛰️ Satellite analysis: Detect disasters with limited labeled geography.
- 🏭 Industrial IoT: Train defect detectors on small sensor datasets.
If you’re Interested in segmentation Model, you may also find this article helpful: 3 Breakthroughs in RGBD Segmentation: How CroDiNo-KD Revolutionizes AI Amid Sensor Failures
The Future: Beyond Image Classification
ActiveKD’s framework extends to:
- 🎥 Video action recognition (tested on UCF101).
- 🧠 Multimodal chatbots (integrating LLaVA/FLAN).
- ⚠️ Limitation: Currently vision-only. Text/audio support is coming!
Conclusion: Stop Wasting 90% of Your AI Budget
ActiveKD and PCoreSet prove you don’t need petabytes of labeled data to train state-of-the-art AI. By combining:
- Zero-shot VLMs as teachers,
- Probability-space active learning,
- Structured bias as a guide—not noise,
You can achieve higher accuracy with 90% fewer labels. The era of “data starvation” is over.
🚀 Call to Action
- Try the code: GitHub Repo (ActiveKD-PCoreSet)
- Read the paper: arXiv: PCoreSet: Effective Active Learning via Knowledge Distillation
- Question for you: Where would YOU deploy ActiveKD first? Comment below! 👇
“The biggest AI cost isn’t compute—it’s annotations. ActiveKD finally cracks this.”
— Lead Researcher, KAIST
Here’s a simplified implementation of the ActiveKD framework with PCoreSet selection based on the paper’s methodology.
# ActiveKD + PCoreSet Implementation
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import DataLoader
from torchvision import models
import numpy as np
from typing import List, Tuple
# ----------------------------
# Vision-Language Teacher Wrapper (CLIP-like)
# ----------------------------
class VisionLanguageTeacher:
def __init__(self, image_encoder, text_encoder, class_prompts, temperature=1.0):
self.image_encoder = image_encoder # f_X
self.text_encoder = text_encoder # f_T
self.class_prompts = class_prompts # ["a photo of a cat", "a photo of a dog", ...]
self.temperature = temperature
self.text_features = self.encode_text(class_prompts)
def encode_text(self, prompts):
with torch.no_grad():
return F.normalize(self.text_encoder(prompts), dim=-1) # (C, d)
def predict(self, images):
with torch.no_grad():
image_features = F.normalize(self.image_encoder(images), dim=-1) # (B, d)
logits = torch.matmul(image_features, self.text_features.T) / self.temperature
probs = F.softmax(logits, dim=-1)
return probs
# ----------------------------
# Student Model Wrapper (e.g., ResNet)
# ----------------------------
class StudentModel(nn.Module):
def __init__(self, base_model, num_classes):
super().__init__()
self.backbone = nn.Sequential(*list(base_model.children())[:-1]) # remove classifier
self.classifier = nn.Linear(base_model.fc.in_features, num_classes)
def forward(self, x):
features = self.backbone(x).squeeze()
return self.classifier(features)
def predict_proba(self, x):
return F.softmax(self.forward(x), dim=-1)
# ----------------------------
# Loss Functions
# ----------------------------
def cross_entropy_loss(pred, label):
return F.cross_entropy(pred, label)
def distillation_loss(student_probs, teacher_probs):
return F.kl_div(student_probs.log(), teacher_probs, reduction='batchmean')
# ----------------------------
# ActiveKD Trainer
# ----------------------------
def train_activekd(student, teacher, labeled_loader, unlabeled_loader, optimizer, lambda_ce=0.5):
student.train()
total_loss = 0
for (x_l, y_l), (x_u,) in zip(labeled_loader, unlabeled_loader):
x_l, y_l, x_u = x_l.cuda(), y_l.cuda(), x_u.cuda()
# Forward
pred_l = student(x_l)
probs_u_student = student.predict_proba(x_u)
probs_u_teacher = teacher.predict(x_u).detach()
# Loss
ce = cross_entropy_loss(pred_l, y_l)
kd = distillation_loss(probs_u_student, probs_u_teacher)
loss = lambda_ce * ce + (1 - lambda_ce) * kd
# Optimize
optimizer.zero_grad()
loss.backward()
optimizer.step()
total_loss += loss.item()
return total_loss
# ----------------------------
# PCoreSet Selection
# ----------------------------
def pcoreset_selection(student, labeled_set, unlabeled_set, query_size):
with torch.no_grad():
labeled_probs = [student.predict_proba(x.unsqueeze(0).cuda()).cpu() for x, _ in labeled_set]
labeled_probs = torch.cat(labeled_probs)
distances = []
for x_u in unlabeled_set:
p_u = student.predict_proba(x_u.unsqueeze(0).cuda()).cpu()
dists = torch.norm(labeled_probs - p_u, dim=1)
distances.append(dists.min().item())
# Select top-k most distant
selected_indices = np.argsort(distances)[-query_size:]
return selected_indices
# ----------------------------
# Example Usage Loop
# ----------------------------
def active_learning_loop(model, teacher, dataset, initial_labeled_idx, rounds=10, query_size=10):
labeled_idx = initial_labeled_idx
unlabeled_idx = list(set(range(len(dataset))) - set(labeled_idx))
for r in range(rounds):
print(f"Round {r+1}/{rounds}")
# Dataloaders
labeled_loader = DataLoader([dataset[i] for i in labeled_idx], batch_size=32, shuffle=True)
unlabeled_loader = DataLoader([dataset[i][0] for i in unlabeled_idx], batch_size=32, shuffle=True)
# Optimizer
optimizer = torch.optim.Adam(model.parameters(), lr=1e-3)
train_activekd(model, teacher, labeled_loader, unlabeled_loader, optimizer)
# PCoreSet Selection
selected = pcoreset_selection(model, [dataset[i] for i in labeled_idx], [dataset[i][0] for i in unlabeled_idx], query_size)
new_indices = [unlabeled_idx[i] for i in selected]
# Update indices
labeled_idx += new_indices
unlabeled_idx = list(set(unlabeled_idx) - set(new_indices))
return model

Pingback: Delayed-KD: A Powerful Breakthrough in Low-Latency Streaming ASR (With a 9.4% CER Reduction) - aitrendblend.com