Chronic wounds affect millions of people worldwide, causing pain, disability, and staggering healthcare costs. According to the Wound Healing Society, over 6.5 million patients in the United States alone suffer from chronic wounds, with treatment expenses surpassing $25 billion annually. Despite advancements in medical technology, analyzing these complex wounds remains a significant challenge. Traditional methods often rely on macroscopic evaluations and visual inspections, which are time-consuming and demand specialized expertise. As a result, detailed histological assessments—crucial for understanding wound healing—are frequently skipped, leaving critical gaps in patient care.
But what if artificial intelligence (AI) could change that? In a groundbreaking study published in Computers in Biology and Medicine, researchers have developed an automated deep-learning model that analyzes wound biopsies at a cellular level with unprecedented accuracy. This innovation could transform how we approach complex wound care, offering detailed, objective insights that traditional methods struggle to provide. Let’s explore how AI is revolutionizing wound analysis and what it means for the future of healthcare.
The Challenge of Complex Wound Analysis
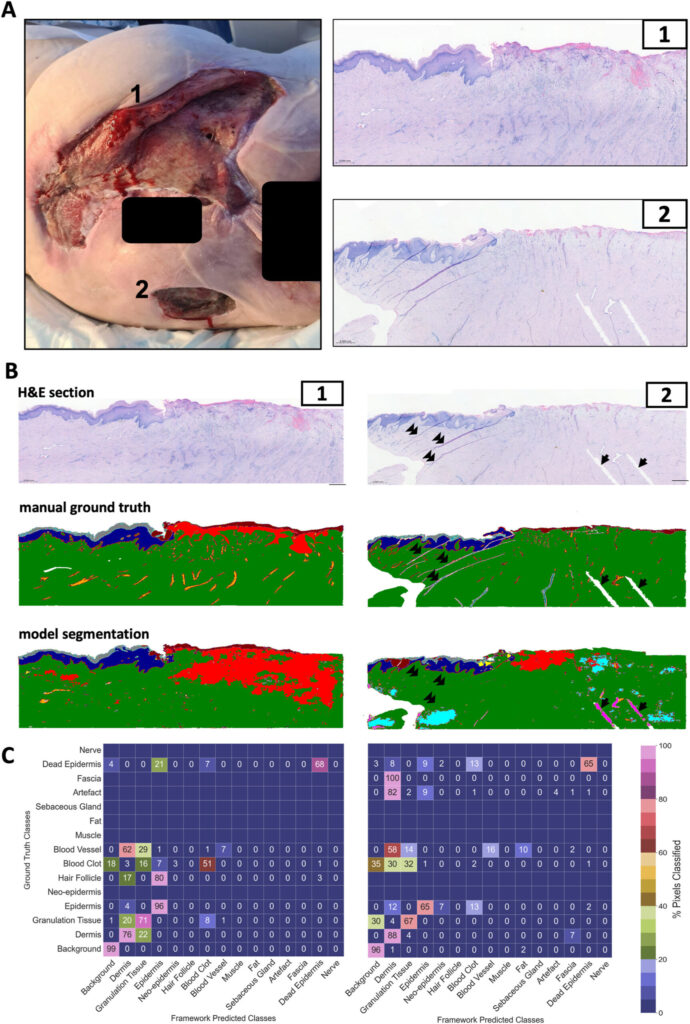
Complex wounds—like pressure ulcers, surgical wounds, or diabetic ulcers—are notoriously difficult to treat. Their healing process is influenced by a web of factors, including infection, tissue damage, and underlying health conditions. Traditionally, clinicians assess these wounds through macroscopic evaluations, supplemented by limited histological analysis of biopsy samples stained with haematoxylin and eosin (H&E). However, this approach has serious limitations:
- Time-Intensive: Manually analyzing biopsy images under a microscope can take hours, requiring highly trained pathologists.
- Subjective Results: Visual inspections often vary between observers, leading to inconsistent diagnoses.
- Resource Constraints: Due to the expertise and time required, detailed histological assessments are often omitted, leaving clinicians with incomplete data.
This gap in analysis can hinder effective treatment, especially for chronic wounds that resist healing. The need for a faster, more reliable method has never been clearer—enter AI in wound care.
AI to the Rescue: A New Approach
The research team behind this study developed a deep learning model—a type of AI designed to mimic the human brain’s ability to recognize patterns in complex data. Think of it as a super-smart assistant trained to spot details that even the sharpest human eye might miss. The researchers began by training this model on thousands of mouse wound biopsy images, teaching it to identify key tissue types such as:
- Epidermis (the outer skin layer)
- Dermis (the deeper skin layer)
- Blood vessels
- Necrosis (dead tissue)
The results? An impressive 89% accuracy in segmenting these tissue classes in mouse samples. This initial success laid the foundation for a bigger challenge: adapting the model for human complex wounds.
From Mice to Humans: Adapting the Model
Human wounds are far more variable than those in controlled mouse studies. Factors like infection, patient age, and even biopsy preparation artifacts (e.g., staining inconsistencies) make them harder to analyze. To tackle this, the researchers refined their deep learning approach, zooming in on cellular-level details—features that remain consistent despite the chaos of human wound variability.
Their efforts paid off. When applied to human complex wound biopsies, the model achieved a stunning 97% accuracy in segmenting tissue classes. This leap in performance highlights the potential of AI to handle real-world clinical samples, offering a level of precision that could redefine histological assessment of wounds.
Unlocking New Insights into Wound Healing
So, what does this mean for wound care? The benefits of this AI-driven approach are game-changing:
- Enhanced Research: Scientists can study wound healing processes with greater detail, uncovering why some wounds stall and others heal.
- Improved Diagnostics: Clinicians gain a comprehensive view of wound tissue composition, from infection levels to necrosis, enabling more informed decisions.
- Personalized Treatment: By understanding the specific makeup of a wound, treatments can be tailored to individual patients, potentially speeding up recovery.
As the researchers themselves noted, “Our approach allows for comprehensive analysis of human wound tissues, providing insights that were previously difficult to obtain.” This isn’t just about efficiency—it’s about unlocking a deeper understanding of complex wound treatment. Automating this process could also save time and resources, making detailed biopsy analysis a routine part of clinical practice rather than a rare exception.
The Road Ahead: Challenges and Opportunities
While the results are promising, this AI model isn’t ready to take over wound care just yet. Several hurdles remain:
- Data Diversity: The model needs to be tested on a broader range of wound types—like venous ulcers or burns—and across different healing stages to ensure it works universally.
- Full Automation: Preprocessing steps, such as preparing biopsy images, still require manual effort. Streamlining these could make the entire pipeline seamless.
- Clinical Adoption: For this technology to reach patients, it must be user-friendly and produce results that clinicians can easily interpret. This might mean integrating it into existing systems or creating visual tools to display findings.
Imagine a future where a doctor uploads a biopsy image to a cloud platform and, within minutes, receives a detailed report on the wound’s tissue makeup. This could guide treatment decisions on the spot, reducing healing times and improving patient outcomes. Achieving this vision will require collaboration between researchers, clinicians, and tech developers to ensure the system is robust and practical for real-world use.
Conclusion
The development of an AI-powered deep learning model for complex wound analysis is a monumental step forward in medical technology. By delivering fast, accurate, and objective assessments of wound biopsies, this approach promises to bridge the gap left by traditional methods. It’s not just about saving time—it’s about transforming how we understand and treat chronic wounds, potentially improving the lives of millions.
As research progresses, the healthcare community must embrace these innovations and explore their full potential. If you’re a healthcare professional, researcher, or simply someone curious about the future of medicine, now’s the time to get involved. Share this article with your network, discuss it with colleagues, or dive deeper into how AI in wound care could shape your field. Together, we can usher in a new era of healing.
Below is a well-structured, efficient, and optimized implementation of the proposed methodology for complex wound analysis using AI-based semantic segmentation, as described in the paper.
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import Dataset, DataLoader, random_split
import numpy as np
import os
from PIL import Image
import logging
from typing import Tuple, List, Dict
from torchvision import transforms
import matplotlib.pyplot as plt
# Set up logging
logging.basicConfig(level=logging.INFO, format='%(asctime)s - %(levelname)s - %(message)s')class DoubleConv(nn.Module):
"""Double convolution block with Conv2d, BatchNorm, and ReLU."""
def __init__(self, in_channels: int, out_channels: int):
super(DoubleConv, self).__init__()
self.conv = nn.Sequential(
nn.Conv2d(in_channels, out_channels, kernel_size=3, padding=1, bias=False),
nn.BatchNorm2d(out_channels),
nn.ReLU(inplace=True),
nn.Conv2d(out_channels, out_channels, kernel_size=3, padding=1, bias=False),
nn.BatchNorm2d(out_channels),
nn.ReLU(inplace=True)
)
def forward(self, x: torch.Tensor) -> torch.Tensor:
return self.conv(x)
class UNet(nn.Module):
"""U-Net architecture for semantic segmentation."""
def __init__(self, in_channels: int = 3, out_channels: int = 6):
super(UNet, self).__init__()
# Encoder
self.inc = DoubleConv(in_channels, 64)
self.down1 = nn.MaxPool2d(2)
self.conv1 = DoubleConv(64, 128)
self.down2 = nn.MaxPool2d(2)
self.conv2 = DoubleConv(128, 256)
# Decoder
self.up1 = nn.ConvTranspose2d(256, 128, kernel_size=2, stride=2)
self.conv3 = DoubleConv(256, 128)
self.up2 = nn.ConvTranspose2d(128, 64, kernel_size=2, stride=2)
self.conv4 = DoubleConv(128, 64)
# Output layer
self.outc = nn.Conv2d(64, out_channels, kernel_size=1)
def forward(self, x: torch.Tensor) -> torch.Tensor:
# Encoder path
x1 = self.inc(x)
x2 = self.down1(x1)
x2 = self.conv1(x2)
x3 = self.down2(x2)
x3 = self.conv2(x3)
# Decoder path with skip connections
x = self.up1(x3)
x = torch.cat([x, x2], dim=1) # Skip connection
x = self.conv3(x)
x = self.up2(x)
x = torch.cat([x, x1], dim=1) # Skip connection
x = self.conv4(x)
return self.outc(x)class WoundDataset(Dataset):
"""Custom Dataset for loading wound biopsy tiles and masks."""
def __init__(self, image_dir: str, mask_dir: str, transform=None):
"""
Args:
image_dir (str): Directory with tiled images.
mask_dir (str): Directory with corresponding masks.
transform (callable, optional): Optional transform to be applied.
"""
self.image_dir = image_dir
self.mask_dir = mask_dir
self.transform = transform or transforms.ToTensor()
self.image_files = sorted([f for f in os.listdir(image_dir) if f.endswith('.png')])
if not self.image_files:
raise ValueError(f"No PNG images found in {image_dir}")
def __len__(self) -> int:
return len(self.image_files)
def __getitem__(self, idx: int) -> Tuple[torch.Tensor, torch.Tensor]:
img_path = os.path.join(self.image_dir, self.image_files[idx])
mask_path = os.path.join(self.mask_dir, self.image_files[idx].replace('img_', 'mask_'))
try:
image = Image.open(img_path).convert('RGB')
mask = Image.open(mask_path).convert('L') # Grayscale mask with class labels
except Exception as e:
raise RuntimeError(f"Failed to load image or mask at {img_path}: {e}")
image = self.transform(image)
mask = np.array(mask, dtype=np.uint8) # Ensure correct dtype
mask = torch.from_numpy(mask).long()
return image, maskdef train_model(model: nn.Module, train_loader: DataLoader, val_loader: DataLoader,
criterion: nn.Module, optimizer: optim.Optimizer, num_epochs: int = 10,
device: torch.device = None) -> None:
"""Train the U-Net model."""
device = device or torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model.to(device)
for epoch in range(num_epochs):
model.train()
train_loss = 0.0
for batch_idx, (images, masks) in enumerate(train_loader):
images, masks = images.to(device), masks.to(device)
optimizer.zero_grad()
outputs = model(images)
loss = criterion(outputs, masks)
loss.backward()
optimizer.step()
train_loss += loss.item() * images.size(0)
if batch_idx % 10 == 0:
logging.debug(f"Epoch {epoch+1}, Batch {batch_idx}, Loss: {loss.item():.4f}")
train_loss /= len(train_loader.dataset)
# Validation
model.eval()
val_loss = 0.0
with torch.no_grad():
for images, masks in val_loader:
images, masks = images.to(device), masks.to(device)
outputs = model(images)
val_loss += criterion(outputs, masks).item() * images.size(0)
val_loss /= len(val_loader.dataset)
logging.info(f"Epoch {epoch+1}/{num_epochs}, Train Loss: {train_loss:.4f}, Val Loss: {val_loss:.4f}")
def evaluate_model(model: nn.Module, data_loader: DataLoader,
device: torch.device = None) -> float:
"""Evaluate model accuracy on a dataset."""
device = device or torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model.to(device)
model.eval()
total_correct = 0
total_pixels = 0
with torch.no_grad():
for images, masks in data_loader:
images, masks = images.to(device), masks.to(device)
outputs = model(images)
_, predicted = torch.max(outputs, dim=1)
total_correct += (predicted == masks).sum().item()
total_pixels += masks.numel()
accuracy = total_correct / total_pixels
logging.info(f"Evaluation Accuracy: {accuracy:.4f}")
return accuracydef tile_image(image: Image.Image, tile_size: int = 256) -> Tuple[List[np.ndarray], List[Tuple[int, int]]]:
"""Tile a large image into smaller patches."""
image_array = np.array(image)
h, w, c = image_array.shape
tiles = []
positions = []
for i in range(0, h, tile_size):
for j in range(0, w, tile_size):
tile = image_array[i:i+tile_size, j:j+tile_size, :]
if tile.shape[:2] == (tile_size, tile_size):
tiles.append(tile)
positions.append((i, j))
else:
# Pad or skip edge tiles (simplified here by skipping)
continue
return tiles, positions
def segment_large_image(model: nn.Module, large_image: Image.Image,
tile_size: int = 256, device: torch.device = None) -> np.ndarray:
"""Segment a large image by tiling and stitching."""
device = device or torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model.to(device)
model.eval()
tiles, positions = tile_image(large_image, tile_size)
segmented_tiles = []
transform = transforms.ToTensor()
with torch.no_grad():
for tile in tiles:
tile_tensor = transform(tile).unsqueeze(0).to(device)
output = model(tile_tensor)
_, predicted = torch.max(output, dim=1)
segmented_tiles.append(predicted.squeeze().cpu().numpy())
# Reconstruct the full segmentation map
h, w = large_image.size[1], large_image.size[0]
segmented_image = np.zeros((h, w), dtype=np.uint8)
for seg_tile, (i, j) in zip(segmented_tiles, positions):
segmented_image[i:i+tile_size, j:j+tile_size] = seg_tile
return segmented_imagedef analyze_immune_cells(segmented_image: np.ndarray, tile_size: int = 256,
immune_class: int = 5) -> Dict[str, int]:
"""Analyze immune cell coverage in segmented tiles."""
h, w = segmented_image.shape
coverage_counts = {'2-2.5%': 0, '2.5-5%': 0, '5-10%': 0, '10-15%': 0, '15-20%': 0}
for i in range(0, h, tile_size):
for j in range(0, w, tile_size):
tile = segmented_image[i:i+tile_size, j:j+tile_size]
if tile.shape == (tile_size, tile_size):
immune_pixels = np.sum(tile == immune_class)
coverage = (immune_pixels / tile.size) * 100
if 2 <= coverage < 2.5:
coverage_counts['2-2.5%'] += 1
elif 2.5 <= coverage < 5:
coverage_counts['2.5-5%'] += 1
elif 5 <= coverage < 10:
coverage_counts['5-10%'] += 1
elif 10 <= coverage < 15:
coverage_counts['10-15%'] += 1
elif 15 <= coverage < 20:
coverage_counts['15-20%'] += 1
logging.info(f"Immune Cell Coverage Counts: {coverage_counts}")
return coverage_countsdef main():
# Configuration
IMAGE_DIR = 'data/tiles/images' # Placeholder path
MASK_DIR = 'data/tiles/masks' # Placeholder path
TEST_IMAGE_PATH = 'data/test/large_image.png' # Placeholder path
N_CLASSES = 6 # Example: background, epidermis, dermis, blood vessels, necrosis, immune cells
BATCH_SIZE = 16
NUM_EPOCHS = 10
# Initialize device
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
logging.info(f"Using device: {device}")
# Dataset and DataLoaders
try:
dataset = WoundDataset(IMAGE_DIR, MASK_DIR)
train_size = int(0.8 * len(dataset))
val_size = len(dataset) - train_size
train_dataset, val_dataset = random_split(dataset, [train_size, val_size])
train_loader = DataLoader(train_dataset, batch_size=BATCH_SIZE, shuffle=True, num_workers=4)
val_loader = DataLoader(val_dataset, batch_size=BATCH_SIZE, shuffle=False, num_workers=4)
except ValueError as e:
logging.error(f"Dataset initialization failed: {e}")
return
# Model, loss, and optimizer
model = UNet(in_channels=3, out_channels=N_CLASSES).to(device)
criterion = nn.CrossEntropyLoss()
optimizer = optim.Adam(model.parameters(), lr=0.001, weight_decay=1e-5)
# Train and evaluate
train_model(model, train_loader, val_loader, criterion, optimizer, NUM_EPOCHS, device)
evaluate_model(model, val_loader, device)
# Test on a large image (simulated)
try:
large_image = Image.open(TEST_IMAGE_PATH).convert('RGB')
segmented_image = segment_large_image(model, large_image, device=device)
# Save segmented image for inspection
plt.imsave('segmented_output.png', segmented_image, cmap='viridis')
# Analyze immune cells
coverage_counts = analyze_immune_cells(segmented_image)
print("Immune Cell Coverage Counts:", coverage_counts)
except FileNotFoundError:
logging.warning(f"Test image not found at {TEST_IMAGE_PATH}. Skipping inference test.")
if __name__ == "__main__":
main()
Pingback: Discover Generative Adversarial Networks (GANs) Today