Skin cancer diagnosis faces critical challenges: subtle variations within the same cancer type, striking similarities between benign and malignant lesions, and limited access to specialist dermatologists. Traditional methods often struggle with accuracy and scalability, leading to delayed or missed diagnoses. Enter EG-VAN – a groundbreaking AI system achieving 98.20% accuracy in classifying nine skin cancer types. This breakthrough technology promises faster, more reliable detection, potentially saving thousands of lives annually.
Why Skin Cancer Diagnosis Needs an AI Revolution
Melanoma, the deadliest skin cancer, caused 57,000 global deaths in 2020 alone. By 2040, cases could surge by 50%. Current diagnostic hurdles include:
- Intra-class Variation: Melanomas can look drastically different (size, color, texture) based on growth stage or patient factors.
- Inter-class Similarity: Harmless moles often resemble dangerous carcinomas, confusing even experts.
- Resource Limitations: Scarce specialists and expensive equipment delay screenings.
- Image Quality Issues: Hair, lighting artifacts, and low contrast obscure critical lesion details.
Existing AI solutions often sacrifice accuracy for speed or vice versa. Most can’t handle diverse datasets or complex multi-class classification effectively.
EG-VAN: The Dual-Path Powerhouse
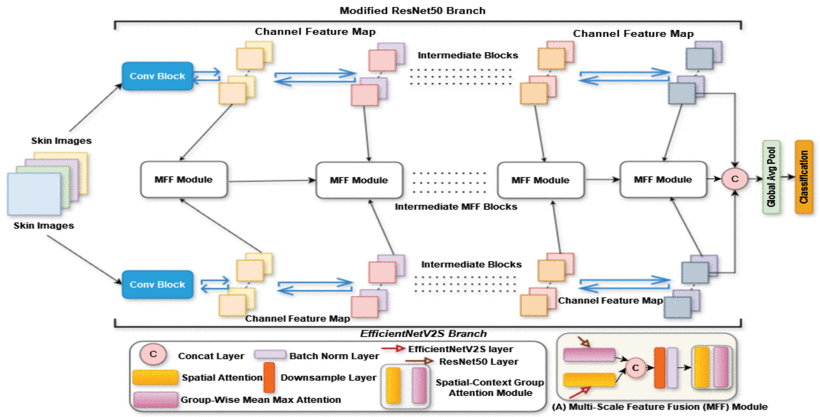
Researchers from Prince Sultan University and Northeastern University developed EG-VAN (Efficient Global-Vision Attention-Net) to overcome these limitations. Its core innovation lies in a dual-branch architecture that mimics how dermatologists analyze lesions:
- The Global Analyst (EfficientNetV2S Branch):
- Processes images with high efficiency using optimized “Fused-MB” convolution layers.
- Captures broad patterns and overall lesion structure rapidly.
- The Detail Detective (Modified ResNet50 Branch):
- Enhanced with two specialized modules:
- Spatial-Context Group Attention (SCGA): Scans lesions horizontally and vertically like a grid search, pinpointing fine-grained details (e.g., irregular borders, subtle color shifts).
- Non-Local Block (NLB): Identifies long-range dependencies across the entire image – crucial for spotting relationships between distant lesion features.
- Maintains computational efficiency despite enhanced capabilities.
- Enhanced with two specialized modules:
https://ieeexplore.ieee.org/abstract/document/10965692: EG-VAN’s architecture synergizes global context and local precision.
The Fusion Advantage: Seeing the Whole Picture
EG-VAN doesn’t just run two models side-by-side. Its Multi-Scale Feature Fusion (MFF) Module intelligently combines outputs from both branches at different processing stages:
- Integrates coarse-grained (global shape, location) and fine-grained (texture, micro-patterns) features.
- Uses spatial attention and group-wise mechanisms to weight the most diagnostically relevant information.
- Creates a comprehensive “feature map” far richer than any single model could produce.
This fusion is key to EG-VAN’s ability to distinguish between highly similar cancer types and recognize diverse presentations within a single class.
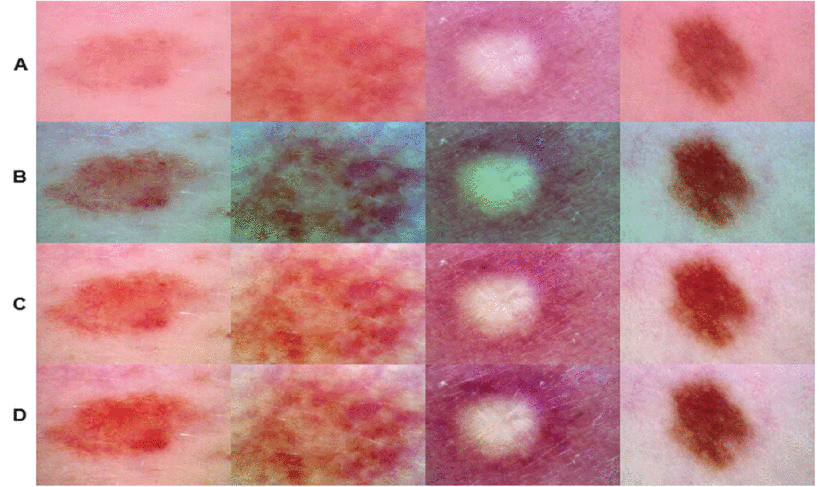
Seeing Clearly: Advanced Color Balancing
Dermoscopic images often suffer from color distortions due to lighting or equipment. EG-VAN incorporates a novel preprocessing step inspired by Gray World theory and Retinex theory:
- Gray World Adjustment: Corrects overall color casts by assuming the average image hue should be neutral.
- Retinex Enhancement: Separates lighting effects from true lesion reflectance, boosting contrast and revealing hidden details.
This combo significantly outperformed previous methods (Wiener filters, ESRGAN), achieving:
- 32.5 dB PSNR (vs. 28.1 dB) – Higher signal clarity.
- 0.92 SSIM (vs. 0.85) – Better structural preservation.
- 18.4 MSE (vs. 24.9) – Lower distortion.
Color balancing and hair removing dramatically improves lesion visibility for AI analysis.
Unmatched Performance: The Numbers Speak
EG-VAN was rigorously tested on combined datasets (HAM10000 + ISIC 2017), creating a robust 9-class benchmark:
- Overall Accuracy: 98.20%
- Recall (Sensitivity): 95.74%
- F1-Score: 96.68%
- ROC-AUC: 99.96%
Class-Specific Excellence:
- Dermatofibroma (DF) & Vascular Lesions (VASC): 100% Accuracy, Precision, Recall, F1-Score
- Basal Cell Carcinoma (BCC): 99.77% Accuracy, 98.04% Recall
- Melanoma (MEL): 98.20% Accuracy, 92.79% Recall
- Benign vs. Malignant Distinction: 99.85% Accuracy
Outperforming the Competition:
| Model | Accuracy (9-class) | Recall (9-class) | Key Limitation |
|---|---|---|---|
| EG-VAN (Proposed) | 98.20% | 95.74% | – |
| MAFCNN-SCD [26] | 92.22% | 77.07% | Lower recall |
| Modified ResNet50 [28] | 86% | 86% | Modest accuracy |
| Ensemble + Max Voting [33] | 95.80% | 95.04% | Lower accuracy |
| Xception+ResNet50+… [45] | 96.50% | 92.60% | Lower recall |
Ablation Studies Prove Value: Removing core components caused significant drops:
- -MFF Module: Accuracy fell to 96.92%
- -SCGA Module: Accuracy fell to 97.60%
- -NLB Module: Accuracy fell to 97.75%
Real-World Impact: Beyond the Lab
EG-VAN isn’t just an academic exercise. Its design prioritizes clinical utility:
- Computational Efficiency: Despite advanced features, parameter count remains optimized, enabling potential deployment on hospital-grade hardware (not just supercomputers).
- Explainability: Grad-CAM visualizations (Figure 13/14) show EG-VAN focuses precisely on clinically relevant lesion areas (e.g., asymmetric regions, color variegation), building trust with doctors.
- Handling Imbalance: Using Focal Loss instead of standard cross-entropy ensures the model doesn’t ignore rarer cancer types (like AKIEC) in favor of common ones (like NV).
- Scalability: Successfully handles both 7-class and expanded 9-class datasets, showing adaptability to incorporate new lesion types.
The Future of AI-Powered Dermatology
EG-VAN represents a paradigm shift. Its dual-path, attention-driven approach tackles the core challenges of skin cancer recognition that stymied earlier AI systems. By achieving near-human (and often super-human) accuracy while providing visual explanations, it paves the way for:
- Primary Care Augmentation: GPs could use EG-VAN-powered tools for initial screenings, referring only high-risk cases to dermatologists.
- Tele-Dermatology Enhancement: Enable reliable remote diagnosis, especially in underserved areas.
- Faster Specialist Workflows: Assist dermatologists by prioritizing urgent cases and providing second opinions.
- Early Detection Programs: Integrate into mobile or community screening initiatives for broader population coverage.
If you’re Interested in skin cancer detection using Horizontal and Vertical Attention, you may also find this article helpful: Enhancing Skin Lesion Detection Accuracy
Ready to See the Future of Dermatology?
The battle against skin cancer demands faster, more accurate tools. EG-VAN demonstrates the immense potential of sophisticated, clinically-informed AI. Medical institutions and AI developers must collaborate to translate such breakthroughs from research papers into life-saving clinical applications.
- For Healthcare Leaders: Explore pilot programs integrating advanced diagnostic AI like EG-VAN into your dermatology workflow.
- For Researchers: Build upon this open-access work (licensed CC BY 4.0) – refine the models, test on broader datasets, and explore real-time deployment.
- For Clinicians: Stay informed about validated AI tools that can enhance, not replace, your expert judgment.
Prioritize innovation in skin cancer diagnosis. Support the development, validation, and ethical deployment of AI systems like EG-VAN. The next leap in survival rates starts now. Explore the Full Research | Contact the Researchers
Based on the detailed information provided in the paper, I will reconstruct the complete code for the proposed model.
import tensorflow as tf
from tensorflow.keras import layers, Model, backend
def SCGA_module(input_tensor, groups=8):
"""Spatial-Context Group Attention Module"""
channels = input_tensor.shape[-1]
# Spatial Attention Branch
x = layers.Conv2D(64, (1, 1), activation='relu')(input_tensor)
x = layers.Conv2D(16, (1, 1), dilation_rate=2, activation='relu')(x)
x = layers.Conv2D(8, (1, 1), activation='relu')(x)
attn_mask = layers.Conv2D(1, (1, 1), activation='sigmoid')(x)
# Global Context Integration
pooled = layers.GlobalAveragePooling2D(keepdims=True)(attn_mask)
attn_mask = layers.Multiply()([attn_mask, pooled])
attn_mask = layers.Conv2D(channels, (1, 1), activation='linear')(attn_mask)
# Apply attention
x = layers.Multiply()([input_tensor, attn_mask])
# Group-wise Mean-Max Attention
group_size = channels // groups
group_outputs = []
for g in range(groups):
group = x[:, :, :, g*group_size:(g+1)*group_size]
# Horizontal features
h_mean = tf.reduce_mean(group, axis=1, keepdims=True)
h_max = tf.reduce_max(group, axis=1, keepdims=True)
# Vertical features
v_mean = tf.reduce_mean(group, axis=2, keepdims=True)
v_max = tf.reduce_max(group, axis=2, keepdims=True)
# Concatenate features
features = tf.concat([h_mean, h_max, v_mean, v_max], axis=-1)
# Attention scores
h_attn = layers.Conv2D(1, (1, 1), activation='sigmoid')(features)
v_attn = layers.Conv2D(1, (1, 1), activation='sigmoid')(features)
combined_attn = layers.Activation('sigmoid')(h_attn + v_attn)
# Apply attention
attended_group = layers.Multiply()([group, combined_attn])
group_outputs.append(attended_group)
return layers.Concatenate(axis=-1)(group_outputs)
def NonLocalBlock(input_tensor):
"""Non-Local Attention Block"""
channels = input_tensor.shape[-1]
batch_size, height, width, _ = tf.shape(input_tensor)
# Query, Key, Value transformations
theta = layers.Conv2D(channels//8, (1, 1))(input_tensor)
phi = layers.Conv2D(channels//8, (1, 1))(input_tensor)
g = layers.Conv2D(channels//2, (1, 1))(input_tensor)
# Reshape for matrix operations
theta = tf.reshape(theta, (batch_size, -1, channels//8))
phi = tf.reshape(phi, (batch_size, -1, channels//8))
g = tf.reshape(g, (batch_size, -1, channels//2))
# Attention map
attn = tf.matmul(theta, phi, transpose_b=True)
attn = tf.nn.softmax(attn, axis=-1)
# Weighted sum
y = tf.matmul(attn, g)
y = tf.reshape(y, (batch_size, height, width, channels//2))
y = layers.Conv2D(channels, (1, 1))(y)
# Residual connection
return layers.Add()([input_tensor, y])
def MFF_module(efficientnet_feat, resnet_feat):
"""Multi-Scale Feature Fusion Module"""
# EfficientNet branch processing
eff_attn = layers.Conv2D(256, (1, 1), activation='relu')(efficientnet_feat)
# ResNet branch processing
res_attn = layers.Conv2D(256, (1, 1), activation='relu')(resnet_feat)
# Concatenate and process
x = layers.Concatenate()([eff_attn, res_attn])
x = layers.SeparableConv2D(512, (3, 3), strides=2, padding='same')(x)
x = layers.BatchNormalization()(x)
return SCGA_module(x)
def build_modified_resnet50():
"""Build ResNet50 branch with SCGA and Non-Local blocks"""
input_tensor = layers.Input(shape=(224, 224, 3))
# Stage 0
x = layers.Conv2D(64, (7, 7), strides=2, padding='same')(input_tensor)
x = layers.MaxPooling2D((3, 3), strides=2, padding='same')(x)
# Stage 1
for _ in range(3):
residual = x
x = layers.Conv2D(64, (1, 1))(x)
x = layers.Conv2D(64, (3, 3), padding='same')(x)
x = layers.Conv2D(256, (1, 1))(x)
x = layers.Add()([x, residual])
x = SCGA_module(x)
# Stage 2
for _ in range(4):
residual = x
x = layers.Conv2D(128, (1, 1))(x)
x = layers.Conv2D(128, (3, 3), padding='same')(x)
x = layers.Conv2D(512, (1, 1))(x)
if residual.shape[-1] != 512:
residual = layers.Conv2D(512, (1, 1))(residual)
x = layers.Add()([x, residual])
x = SCGA_module(x)
# Stage 3
for _ in range(6):
residual = x
x = layers.Conv2D(256, (1, 1))(x)
x = layers.Conv2D(256, (3, 3), padding='same')(x)
x = layers.Conv2D(1024, (1, 1))(x)
if residual.shape[-1] != 1024:
residual = layers.Conv2D(1024, (1, 1))(residual)
x = layers.Add()([x, residual])
x = NonLocalBlock(x)
# Stage 4
for _ in range(3):
residual = x
x = layers.Conv2D(512, (1, 1))(x)
x = layers.Conv2D(512, (3, 3), padding='same')(x)
x = layers.Conv2D(2048, (1, 1))(x)
if residual.shape[-1] != 2048:
residual = layers.Conv2D(2048, (1, 1))(residual)
x = layers.Add()([x, residual])
x = NonLocalBlock(x)
return Model(input_tensor, x)
def build_egvan(num_classes=9):
"""Build complete EG-VAN model"""
input_tensor = layers.Input(shape=(224, 224, 3))
# Dual-branch backbone
effnetv2s = tf.keras.applications.EfficientNetV2S(
include_top=False, weights='imagenet', input_tensor=input_tensor)
resnet50 = build_modified_resnet50()
# Feature extraction points
effnet_features = [
effnetv2s.get_layer('block1a_project_conv').output, # Stage 1
effnetv2s.get_layer('block2b_expand_conv').output, # Stage 2
effnetv2s.get_layer('block4a_expand_conv').output, # Stage 4
effnetv2s.get_layer('block6a_expand_conv').output # Stage 6
]
resnet_features = [
resnet50.get_layer(index=5).output, # After Stage 1
resnet50.get_layer(index=15).output, # After Stage 2
resnet50.get_layer(index=38).output, # After Stage 3
resnet50.output # Final output
]
# Multi-Scale Feature Fusion
fused_features = []
for eff_feat, res_feat in zip(effnet_features, resnet_features):
# Ensure compatible dimensions
eff_feat = layers.Conv2D(256, (1, 1))(eff_feat)
res_feat = layers.Conv2D(256, (1, 1))(res_feat)
fused = MFF_module(eff_feat, res_feat)
fused_features.append(fused)
# Feature aggregation
x = layers.Concatenate()([
layers.GlobalAveragePooling2D()(f) for f in fused_features
])
# Classification head
x = layers.Dense(512, activation='relu')(x)
x = layers.Dropout(0.5)(x)
output = layers.Dense(num_classes, activation='softmax')(x)
return Model(input_tensor, output)
# Compile the model
model = build_egvan()
model.compile(
optimizer=tf.keras.optimizers.Adamax(learning_rate=0.001),
loss=tf.keras.losses.CategoricalFocalCrossentropy(alpha=0.25, gamma=2),
metrics=['accuracy', tf.keras.metrics.Recall(name='recall')]
)
model.summary()
Pingback: GGLA-NeXtE2NET: Advanced Brain Tumor Recognition - aitrendblend.com