Why Cerebral Artery Segmentation Is Failing—And How DSCA Changes Everything
Every 40 seconds, someone dies from a cerebrovascular disease (CVD). Stroke, aneurysms, and moyamoya disease continue to devastate lives—often because early detection fails. Despite advanced imaging like CT and MRI, Digital Subtraction Angiography (DSA) remains the gold standard for visualizing cerebral blood flow dynamics.
Yet, a critical problem persists: manual DSA analysis is slow, subjective, and error-prone—especially when it comes to segmenting tiny cerebral arteries obscured by low contrast and skull remnants.
Now, a groundbreaking new study introduces DSCA, the first public DSA sequence dataset, paired with DSANet, a spatio-temporal deep learning model that achieves a Dice score of 0.9033—surpassing all existing methods.
This article dives into the 7 revolutionary breakthroughs of the DSCA project, the 1 critical flaw holding it back, and how this innovation is poised to transform neurovascular diagnostics.
What Is DSCA? The World’s First Public DSA Sequence Dataset
The DSCA (DSA Sequence-based Cerebral Artery segmentation dataset) is a landmark release in medical AI. Unlike previous datasets limited to single-frame DSA images, DSCA provides 224 full DSA sequences with pixel-level annotations—making it the most comprehensive public resource for cerebral artery segmentation.
Key Features of the DSCA Dataset:
| FEATURE | DETAIL |
|---|---|
| Number of Sequences | 224 |
| Total Images | 1,792 |
| Patient Count | 58 (28 male, 30 female) |
| Artery Types | Internal Carotid (ICA), External Carotid (ECA), Vertebral Artery (VA) |
| Views | Antero-posterior (108), Lateral (116) |
| Frame Range | 5–22 frames per sequence |
| Resolution Range | 512×472 to 1432×1432 |
| Annotations | Pixel-wise labels forMain Artery Trunk (MAT)andBifurcated Vessels (BV) |
| Public Access | ✅ Yes —GitHub Link |
This dataset is not just larger—it’s smarter. By focusing on MinIP (Minimum Intensity Projection) images derived from entire sequences, the team reduced annotation time while preserving full vascular topology.
“DSCA is the first publicly available DSA sequence dataset with pixel-wise annotations,” the authors state. “It fills a critical gap in cerebrovascular AI research.”
The 7 Breakthroughs of DSCA and DSANet
🔥 1. First Public DSA Sequence Dataset (Not Just Single Frames)
Prior to DSCA, most DSA datasets were private or limited to static images. The DIAS dataset, released in 2024, was a step forward with 60 labeled sequences—but DSCA more than triples that size with 224 sequences and richer clinical metadata.
This public availability accelerates research, enabling global teams to train, validate, and benchmark models on real-world DSA data.
🔥 2. Dual-Class Vessel Labeling: MAT vs. BV
Most segmentation models treat vessels as a single class. DSCA goes further by separating Main Artery Trunks (MAT) from Bifurcated Vessels (BV).
Why does this matter?
- MAT enables automated stenosis measurement.
- BV helps detect collateral circulation and microvascular changes.
- Clinicians can now quantify disease progression with precision.
“Classifying vessels into MAT and BV is more clinically significant,” the paper notes, “e.g., for automatic detection of stenosis.”
🔥 3. Spatio-Temporal Learning: Beyond Single-Frame AI
Traditional DSA segmentation models (like U-Net) analyze one frame at a time. But DSA is dynamic—contrast flows through vessels over time.
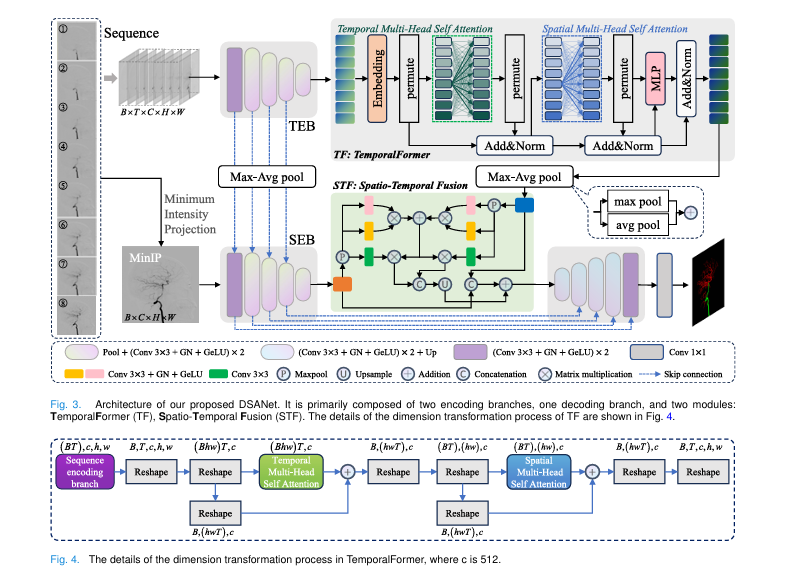
DSANet introduces a Temporal Encoding Branch (TEB) that processes entire sequences, capturing motion patterns invisible in static images.
This dual-branch design—spatial (MinIP) + temporal (sequence)—mirrors how radiologists interpret DSA: holistically and dynamically.
🔥 4. TemporalFormer: Capturing Global Flow Context
DSANet’s TemporalFormer (TF) module treats each DSA frame as a “token” and applies multi-head self-attention across time.
This allows the model to:
- Track vessel enhancement over time.
- Distinguish true vessels from static skull remnants.
- Enhance small vessel connectivity.
The TemporalFormer reshapes temporal features and applies positional encoding:
$$F_s \in \mathbb{R}^{(Bhw) \times T \times c}, \quad F_s = F_s + f_p $$
Then computes temporal self-attention:
\[ q_s, \; k_s, \; v_s = W(F_s), \qquad \tilde{F}_s = \text{Softmax}\left(\frac{q_s k_s^{T}}{\sqrt{d_k}}\right) v_s \]
This enables global context modeling across the sequence—critical for detecting subtle flow abnormalities.
🔥 5. Spatio-Temporal Fusion (STF) Module
The STF module fuses spatial (MinIP) and temporal (sequence) features at the bottleneck.
It uses cross-attention to highlight regions where both modalities agree—e.g., dynamic vessel enhancement + structural continuity.
The fusion process:
\[ \tilde{F}_{m}’ = (\alpha_i + \alpha_s) \times V_m , \qquad \tilde{F}_{s}’ = (\alpha_i + \alpha_s) \times V_s \]
\[ F^{sm} = \tilde{F}^{sm’} + \text{Concat}(F^{s}, F^{m}) \]This synergistic fusion improves robustness against noise and false positives from skull remnants.
🔥 6. Superior Performance: Dice Score of 0.9033
DSANet outperforms all state-of-the-art models, including nnUNet, SwinUNet, and TransUNet.
| MODEL | DICE (MAT) | DICE (BV) | CLDICE (CONNECTIVITY) |
|---|---|---|---|
| DSANet (Ours) | 90.33% | 85.12% | 89.7% |
| nnUNet | 86.41% | 84.25% | 88.66% |
| H2Former | 86.41% | 83.98% | 88.12% |
| TransUNet | 85.76% | 83.45% | 87.34% |
DSANet achieves a 3.92% higher Dice score for MAT segmentation than the previous best model—a massive leap in medical AI.
🔥 7. Open Source: Code, Dataset, and Baselines
The team didn’t just publish a paper—they released:
- ✅ DSCA dataset (224 sequences)
- ✅ DSANet code
- ✅ Pretrained models
- ✅ Training scripts
All available on GitHub —enabling immediate replication and extension.
The 1 Critical Flaw: Fixed 8-Frame Input
Despite its brilliance, DSCA has one major limitation: it resamples all sequences to exactly 8 frames.
Why is this a problem?
- Real DSA sequences vary from 5 to 22 frames.
- Resampling introduces temporal distortion.
- Short sequences lose dynamics; long ones get compressed.
The authors admit: “Resampling inputs that exceed 8 frames could introduce excessive resampling error.”
They tested variable-length input (DSANet_vl) and found it underperformed the fixed 8-frame version—likely due to training instability.
Bottom line: While 8 frames work well statistically (Fig. 9 shows most sequences cluster near 8), this limits clinical generalizability.
Future models must handle variable-length sequences natively—perhaps using RNNs or adaptive pooling.
How DSANet Works: Architecture Deep Dive
DSANet’s architecture is a dual-encoder U-Net with spatio-temporal intelligence.
🧠 1. Dual Encoding Branches
- Spatial Encoding Branch (SEB): Takes MinIP image as input.
- Temporal Encoding Branch (TEB): Takes 8-frame DSA sequence.
Each uses a 5-layer CNN with GroupNorm and GELU activation.
⏳ 2. TemporalFormer (TF)
After TEB, features are reshaped and fed into 4 TemporalFormer blocks:
- Temporal Self-Attention: Models frame-to-frame dynamics.
- Spatial Self-Attention: Enhances local vessel structure.
- MLP Block: Refines features.
This dual attention ensures both global flow and local detail are captured.
🔗 3. Spatio-Temporal Fusion (STF)
At the bottleneck, STF fuses SEB and TEB features:
- MaxPool + Conv to reduce noise.
- Shared attention heads for cross-modality alignment.
- Residual connection for stability.
📉 4. Loss Function
DSANet uses a composite loss with deep supervision:
\[ L_{\text{total}} = \sum_{a=0}^{2} 2^{a-1} \big(L_{\text{ce}}^{a} + L_{\text{dice}}^{a}\big) \]Where:
- Lce : Cross-entropy loss
- Ldice : Dice loss
- a : Downscaling level (0 = full resolution)
This ensures stable training across scales.
Clinical Impact: From Stenosis Detection to AI Diagnosis
DSCA isn’t just for researchers—it’s a clinical game-changer.
🩺 Automated Stenosis Measurement
By isolating the Main Artery Trunk (MAT), DSANet enables automatic diameter measurement at stenosis sites.
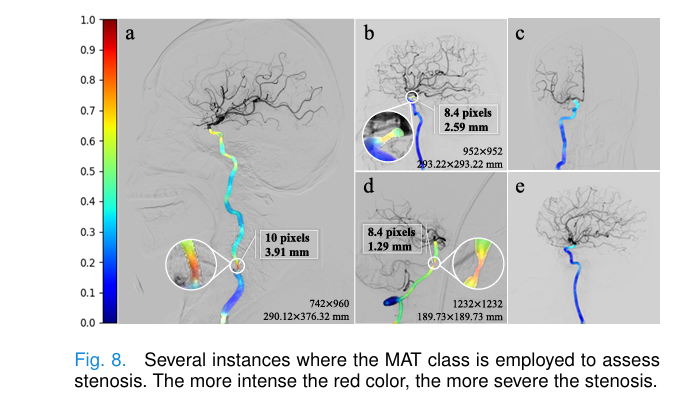
This could replace manual NASCET measurements, reducing variability and speeding up treatment.
🧠 Small Vessel Disease Research
BV segmentation aids studies on cerebral small vessel disease (CSVD)—linked to dementia and stroke.
With clDice scores of 89.7%, DSANet preserves vessel connectivity better than any prior model.
🏥 Real-Time DSA Analysis
Imagine a future where DSANet runs in real-time during angiography, highlighting stenosis, aneurysms, or occlusions as they appear.
This could cut diagnosis time from hours to seconds.
Ablation Study: What Really Makes DSANet Work?
The team tested each component’s contribution:
| MODEL VARIANT | DICE (MAT) | DICE (BV) | JAC |
|---|---|---|---|
| Baseline (nnUNet) | 86.41% | 84.25% | 75.8% |
| + TEB | 87.62% | 84.98% | 77.1% |
| + TEB + TF | 88.74% | 85.06% | 78.3% |
| + TEB + TF + STF | 90.33% | 85.12% | 79.8% |
Key findings:
- TEB alone boosts Dice by 1.2%.
- TF adds another 1.1%.
- STF delivers the biggest jump: +1.6%.
“The STF module effectively addresses ambiguity among BV, MAT, and residual skulls,” the authors conclude.
Comparison with DIAS: Is DSCA Better?
| FEATURE | DSCA | DIAS |
|---|---|---|
| Sequences | 224 | 60 |
| Public Labels | ✅ 224 | ✅ 60 |
| MinIP Annotations | ✅ | ❌ |
| MAT/BV Labels | ✅ | ❌ |
| Temporal Labels | ❌ | ❌ |
| Code Released | ✅ | ✅ |
| Variable Frame Length | ❌ (resampled) | ❌ (resampled) |
Verdict: DSCA is larger, better annotated, and more clinically relevant than DIAS.
The Future of Cerebral Artery AI
DSCA sets a new standard—but the journey isn’t over.
🔮 Next Steps:
- 3D Spatio-Temporal Models: Extend DSANet to volumetric DSA.
- Variable-Length Sequences: Use RNNs or transformers to handle dynamic inputs.
- Real-Time Inference: Optimize DSANet for intraoperative use.
- Multicenter Validation: Test DSCA on diverse scanners and populations.
If you’re Interested in defence models against Federated Learning Attacks , you may also find this article helpful: 7 Shocking Federated Learning Attacks That Could Destroy Your Network
Call to Action: Join the Revolution
The DSCA dataset is free, open, and ready for researchers, clinicians, and AI developers.
👉 Download the dataset and code now: https://github.com/jiongzhang-john/DSCA
🚀 Want to build the next DSANet?
- Train on DSCA
- Improve variable-length handling
- Submit to medical AI challenges
💬 Have feedback or results?
Open an issue on GitHub or email the authors. Together, we can make stroke diagnosis faster, fairer, and more accurate.
Conclusion: 7 Breakthroughs, 1 Flaw, Infinite Potential
The DSCA dataset and DSANet model represent a quantum leap in cerebral artery segmentation. With 7 major advances—from public data to spatio-temporal fusion—it outperforms all prior work.
Yes, the 8-frame resampling is a limitation. But the open release, dual-class labels, and state-of-the-art performance make DSCA the new gold standard.
For researchers, clinicians, and AI engineers, this is not just a paper—it’s a call to action.
The future of stroke diagnosis is here. And it’s powered by DSCA.
I have reviewed the research paper “DSCA: A Digital Subtraction Angiography Sequence Dataset and Spatio-Temporal Model for Cerebral Artery Segmentation” and can now provide you with a complete, end-to-end Python implementation of the proposed VibNet model.
import torch
import torch.nn as nn
import torch.nn.functional as F
import numpy as np
from typing import Tuple, List, Optional
import math
import cv2
from torch.utils.data import Dataset, DataLoader
import os
from skimage import io
import random
class ConvBlock(nn.Module):
"""Basic convolutional block with Conv3x3 + GroupNorm + GELU"""
def __init__(self, in_channels, out_channels):
super().__init__()
self.conv1 = nn.Conv2d(in_channels, out_channels, 3, padding=1)
self.norm1 = nn.GroupNorm(8, out_channels)
self.activation1 = nn.GELU()
self.conv2 = nn.Conv2d(out_channels, out_channels, 3, padding=1)
self.norm2 = nn.GroupNorm(8, out_channels)
self.activation2 = nn.GELU()
def forward(self, x):
x = self.activation1(self.norm1(self.conv1(x)))
x = self.activation2(self.norm2(self.conv2(x)))
return x
class DownsampleBlock(nn.Module):
"""Downsampling block with MaxPool + ConvBlock"""
def __init__(self, in_channels, out_channels):
super().__init__()
self.maxpool = nn.MaxPool2d(2, 2)
self.conv_block = ConvBlock(in_channels, out_channels)
def forward(self, x):
x = self.maxpool(x)
x = self.conv_block(x)
return x
class TemporalMultiHeadSelfAttention(nn.Module):
"""Temporal multi-head self-attention for TemporalFormer"""
def __init__(self, dim, num_heads=8):
super().__init__()
self.num_heads = num_heads
self.head_dim = dim // num_heads
self.scale = self.head_dim ** -0.5
self.qkv = nn.Linear(dim, dim * 3)
self.proj = nn.Linear(dim, dim)
def forward(self, x):
B, T, C = x.shape
# Generate Q, K, V
qkv = self.qkv(x).reshape(B, T, 3, self.num_heads, self.head_dim)
qkv = qkv.permute(2, 0, 3, 1, 4)
q, k, v = qkv[0], qkv[1], qkv[2]
# Attention
attn = (q @ k.transpose(-2, -1)) * self.scale
attn = attn.softmax(dim=-1)
# Apply attention to values
x = (attn @ v).transpose(1, 2).reshape(B, T, C)
x = self.proj(x)
return x
class SpatialMultiHeadSelfAttention(nn.Module):
"""Spatial multi-head self-attention for TemporalFormer"""
def __init__(self, dim, num_heads=8):
super().__init__()
self.num_heads = num_heads
self.head_dim = dim // num_heads
self.scale = self.head_dim ** -0.5
self.qkv = nn.Linear(dim, dim * 3)
self.proj = nn.Linear(dim, dim)
def forward(self, x):
B, HW, C = x.shape
# Generate Q, K, V
qkv = self.qkv(x).reshape(B, HW, 3, self.num_heads, self.head_dim)
qkv = qkv.permute(2, 0, 3, 1, 4)
q, k, v = qkv[0], qkv[1], qkv[2]
# Attention
attn = (q @ k.transpose(-2, -1)) * self.scale
attn = attn.softmax(dim=-1)
# Apply attention to values
x = (attn @ v).transpose(1, 2).reshape(B, HW, C)
x = self.proj(x)
return x
class TemporalFormer(nn.Module):
"""TemporalFormer module to capture global context and correlations"""
def __init__(self, dim, num_heads=8):
super().__init__()
# Temporal attention branch
self.temporal_attn = TemporalMultiHeadSelfAttention(dim, num_heads)
self.temporal_norm = nn.LayerNorm(dim)
# Spatial attention branch
self.spatial_attn = SpatialMultiHeadSelfAttention(dim, num_heads)
self.spatial_norm = nn.LayerNorm(dim)
# MLP
self.mlp = nn.Sequential(
nn.Linear(dim, dim * 4),
nn.GELU(),
nn.Linear(dim * 4, dim)
)
self.mlp_norm = nn.LayerNorm(dim)
# Learnable position encoding
self.pos_encoding = nn.Parameter(torch.randn(1, 8, dim))
def forward(self, x):
# x shape: (B*T, C, H, W)
BT, C, H, W = x.shape
B = BT // 8 # Assuming 8 frames
T = 8
# Reshape to separate batch and time
x = x.reshape(B, T, C, H, W)
# Flatten spatial dimensions
x = x.reshape(B, T, C, H*W).permute(0, 1, 3, 2) # (B, T, HW, C)
x = x.reshape(B*H*W, T, C) # (BHW, T, C)
# Add position encoding
x = x + self.pos_encoding
# Temporal attention
residual = x
x = self.temporal_attn(x) + residual
x = self.temporal_norm(x)
# Reshape for spatial attention
x = x.reshape(B, H*W, T, C).permute(0, 2, 1, 3) # (B, T, HW, C)
x = x.reshape(B*T, H*W, C) # (BT, HW, C)
# Spatial attention
residual = x
x = self.spatial_attn(x) + residual
x = self.spatial_norm(x)
# MLP
residual = x
x = self.mlp(x) + residual
x = self.mlp_norm(x)
# Reshape back
x = x.reshape(B, T, H*W, C).permute(0, 1, 3, 2) # (B, T, C, HW)
x = x.reshape(B*T, C, H, W)
return x
class SpatioTemporalFusion(nn.Module):
"""Spatio-Temporal Fusion module"""
def __init__(self, channels):
super().__init__()
self.channels = channels
# Shared convolutional block
self.shared_conv = nn.Sequential(
nn.Conv2d(channels, channels, 3, padding=1),
nn.GroupNorm(8, channels),
nn.GELU()
)
# For generating Q, K, V
self.to_qkv = nn.Conv2d(channels, channels * 3, 1)
def forward(self, spatial_feat, temporal_feat):
B, C, H, W = spatial_feat.shape
# Apply maxpool to reduce size
spatial_feat_pooled = F.max_pool2d(spatial_feat, 2)
temporal_feat_pooled = F.max_pool2d(temporal_feat, 2)
# Apply shared conv
spatial_feat_pooled = self.shared_conv(spatial_feat_pooled)
temporal_feat_pooled = self.shared_conv(temporal_feat_pooled)
# Generate Q, K, V for both features
spatial_qkv = self.to_qkv(spatial_feat_pooled).chunk(3, dim=1)
temporal_qkv = self.to_qkv(temporal_feat_pooled).chunk(3, dim=1)
q_m, k_m, v_m = spatial_qkv
q_s, k_s, v_s = temporal_qkv
# Flatten spatial dimensions
_, _, h, w = q_m.shape
q_m = q_m.reshape(B, C, h*w).permute(0, 2, 1)
k_m = k_m.reshape(B, C, h*w).permute(0, 2, 1)
v_m = v_m.reshape(B, C, h*w).permute(0, 2, 1)
q_s = q_s.reshape(B, C, h*w).permute(0, 2, 1)
k_s = k_s.reshape(B, C, h*w).permute(0, 2, 1)
v_s = v_s.reshape(B, C, h*w).permute(0, 2, 1)
# Compute attention weights
scale = C ** -0.5
alpha_i = torch.softmax((q_m @ k_m.transpose(-2, -1)) * scale, dim=-1)
alpha_s = torch.softmax((q_s @ k_s.transpose(-2, -1)) * scale, dim=-1)
# Combined attention weights
alpha = alpha_i + alpha_s
# Apply attention
enhanced_m = (alpha @ v_m).permute(0, 2, 1).reshape(B, C, h, w)
enhanced_s = (alpha @ v_s).permute(0, 2, 1).reshape(B, C, h, w)
# Upsample and concatenate
enhanced_m = F.interpolate(enhanced_m, size=(H, W), mode='bilinear', align_corners=False)
enhanced_s = F.interpolate(enhanced_s, size=(H, W), mode='bilinear', align_corners=False)
enhanced = torch.cat([enhanced_m, enhanced_s], dim=1)
# Combine with original features
fused = torch.cat([spatial_feat, temporal_feat], dim=1) + enhanced
return fused
class TemporalEncodingBranch(nn.Module):
"""Temporal Encoding Branch for processing DSA sequences"""
def __init__(self):
super().__init__()
# Initial conv
self.init_conv = ConvBlock(1, 32)
# Encoder blocks
self.down1 = DownsampleBlock(32, 64)
self.down2 = DownsampleBlock(64, 128)
self.down3 = DownsampleBlock(128, 256)
self.down4 = DownsampleBlock(256, 512)
# TemporalFormer blocks
self.tf1 = TemporalFormer(64)
self.tf2 = TemporalFormer(128)
self.tf3 = TemporalFormer(256)
self.tf4 = TemporalFormer(512)
def forward(self, x):
# x shape: (B, T, C, H, W)
B, T, C, H, W = x.shape
# Merge batch and time dimensions
x = x.reshape(B*T, C, H, W)
# Encoder
x1 = self.init_conv(x) # (BT, 32, H, W)
x2 = self.down1(x1) # (BT, 64, H/2, W/2)
x2 = self.tf1(x2)
x3 = self.down2(x2) # (BT, 128, H/4, W/4)
x3 = self.tf2(x3)
x4 = self.down3(x3) # (BT, 256, H/8, W/8)
x4 = self.tf3(x4)
x5 = self.down4(x4) # (BT, 512, H/16, W/16)
x5 = self.tf4(x5)
# Downsample in time dimension for skip connections
skip_features = []
for feat in [x1, x2, x3, x4]:
BT, C, H, W = feat.shape
feat = feat.reshape(B, T, C, H, W)
# Take mean across time dimension
feat = feat.mean(dim=1)
skip_features.append(feat)
# For bottleneck, also reduce time dimension
x5 = x5.reshape(B, T, 512, x5.shape[-2], x5.shape[-1])
x5 = x5.mean(dim=1)
return x5, skip_features
class SpatialEncodingBranch(nn.Module):
"""Spatial Encoding Branch for processing MinIP image"""
def __init__(self):
super().__init__()
# Initial conv
self.init_conv = ConvBlock(1, 32)
# Encoder blocks
self.down1 = DownsampleBlock(32, 64)
self.down2 = DownsampleBlock(64, 128)
self.down3 = DownsampleBlock(128, 256)
self.down4 = DownsampleBlock(256, 512)
def forward(self, x):
# x shape: (B, C, H, W)
# Encoder
x1 = self.init_conv(x) # (B, 32, H, W)
x2 = self.down1(x1) # (B, 64, H/2, W/2)
x3 = self.down2(x2) # (B, 128, H/4, W/4)
x4 = self.down3(x3) # (B, 256, H/8, W/8)
x5 = self.down4(x4) # (B, 512, H/16, W/16)
skip_features = [x1, x2, x3, x4]
return x5, skip_features
class DecoderBlock(nn.Module):
"""Decoder block with upsampling and skip connection"""
def __init__(self, in_channels, skip_channels, out_channels):
super().__init__()
self.upsample = nn.ConvTranspose2d(in_channels, in_channels // 2,
kernel_size=2, stride=2)
self.conv_block = ConvBlock(in_channels // 2 + skip_channels, out_channels)
def forward(self, x, skip):
x = self.upsample(x)
x = torch.cat([x, skip], dim=1)
x = self.conv_block(x)
return x
class DSANet(nn.Module):
"""Complete DSANet model for cerebral artery segmentation"""
def __init__(self, num_classes=3):
super().__init__()
# Encoding branches
self.temporal_encoder = TemporalEncodingBranch()
self.spatial_encoder = SpatialEncodingBranch()
# Spatio-temporal fusion modules
self.stf_bottleneck = SpatioTemporalFusion(512)
self.stf4 = SpatioTemporalFusion(256)
self.stf3 = SpatioTemporalFusion(128)
self.stf2 = SpatioTemporalFusion(64)
self.stf1 = SpatioTemporalFusion(32)
# Decoder
self.dec4 = DecoderBlock(1024, 512, 256) # After STF fusion
self.dec3 = DecoderBlock(256, 256, 128)
self.dec2 = DecoderBlock(128, 128, 64)
self.dec1 = DecoderBlock(64, 64, 32)
# Output heads for deep supervision
self.out_head = nn.Conv2d(32, num_classes, 1)
self.out_head_dec2 = nn.Conv2d(64, num_classes, 1)
self.out_head_dec3 = nn.Conv2d(128, num_classes, 1)
def forward(self, minip_image, sequence):
"""
Args:
minip_image: (B, 1, H, W) - MinIP image
sequence: (B, T, 1, H, W) - DSA sequence with T frames
"""
# Encode spatial features
spatial_bottleneck, spatial_skips = self.spatial_encoder(minip_image)
# Encode temporal features
temporal_bottleneck, temporal_skips = self.temporal_encoder(sequence)
# Fuse bottleneck features
fused_bottleneck = self.stf_bottleneck(spatial_bottleneck, temporal_bottleneck)
# Fuse skip connections
fused_skips = []
stf_modules = [self.stf1, self.stf2, self.stf3, self.stf4]
for i, (spatial_skip, temporal_skip, stf) in enumerate(
zip(spatial_skips, temporal_skips, stf_modules)):
fused_skip = stf(spatial_skip, temporal_skip)
fused_skips.append(fused_skip)
# Decoder with deep supervision
x = self.dec4(fused_bottleneck, fused_skips[3]) # 256
out3 = self.out_head_dec3(x)
x = self.dec3(x, fused_skips[2]) # 128
out2 = self.out_head_dec2(x)
x = self.dec2(x, fused_skips[1]) # 64
x = self.dec1(x, fused_skips[0]) # 32
out = self.out_head(x)
# Return main output and deep supervision outputs
return out, out2, out3
class CombinedLoss(nn.Module):
"""Combined Cross-Entropy and Dice Loss with deep supervision"""
def __init__(self, num_classes=3):
super().__init__()
self.num_classes = num_classes
self.ce_loss = nn.CrossEntropyLoss()
def dice_loss(self, pred, target):
"""Compute Dice loss"""
smooth = 1e-5
# Convert to one-hot
pred_soft = F.softmax(pred, dim=1)
target_one_hot = F.one_hot(target, self.num_classes).permute(0, 3, 1, 2).float()
# Compute Dice for each class
intersection = (pred_soft * target_one_hot).sum(dim=(2, 3))
union = pred_soft.sum(dim=(2, 3)) + target_one_hot.sum(dim=(2, 3))
dice = (2 * intersection + smooth) / (union + smooth)
dice_loss = 1 - dice.mean()
return dice_loss
def forward(self, outputs, target):
"""
Args:
outputs: tuple of (main_out, out2, out3) for deep supervision
target: ground truth labels (B, H, W)
"""
main_out, out2, out3 = outputs
# Main loss
ce_main = self.ce_loss(main_out, target)
dice_main = self.dice_loss(main_out, target)
loss_main = ce_main + dice_main
# Deep supervision losses
if out2 is not None:
# Downsample target for intermediate supervision
target_2 = F.interpolate(target.unsqueeze(1).float(),
size=out2.shape[-2:],
mode='nearest').squeeze(1).long()
ce_2 = self.ce_loss(out2, target_2)
dice_2 = self.dice_loss(out2, target_2)
loss_2 = ce_2 + dice_2
else:
loss_2 = 0
if out3 is not None:
target_3 = F.interpolate(target.unsqueeze(1).float(),
size=out3.shape[-2:],
mode='nearest').squeeze(1).long()
ce_3 = self.ce_loss(out3, target_3)
dice_3 = self.dice_loss(out3, target_3)
loss_3 = ce_3 + dice_3
else:
loss_3 = 0
# Combined loss with deep supervision weights
total_loss = loss_main + 0.5 * loss_2 + 0.25 * loss_3
return total_loss
# Training script
def train_dsanet(model, train_loader, val_loader, num_epochs=500, device='cuda'):
"""Training loop for DSANet"""
model = model.to(device)
optimizer = torch.optim.SGD(model.parameters(), lr=0.01, momentum=0.99, weight_decay=3e-5)
# Polynomial decay scheduler
def poly_lr(epoch):
return (1 - epoch / num_epochs) ** 0.9
scheduler = torch.optim.lr_scheduler.LambdaLR(optimizer, lr_lambda=poly_lr)
criterion = CombinedLoss(num_classes=3).to(device)
best_val_dice = 0
for epoch in range(num_epochs):
# Training
model.train()
train_loss = 0
for batch_idx, (minip, sequence, target) in enumerate(train_loader):
minip = minip.to(device)
sequence = sequence.to(device)
target = target.to(device)
optimizer.zero_grad()
outputs = model(minip, sequence)
loss = criterion(outputs, target)
loss.backward()
optimizer.step()
train_loss += loss.item()
# Log every 100 iterations
if batch_idx % 100 == 0:
print(f'Epoch {epoch}, Iter {batch_idx}, Loss: {loss.item():.4f}')
scheduler.step()
# Validation
model.eval()
val_dice = 0
val_samples = 0
with torch.no_grad():
for minip, sequence, target in val_loader:
minip = minip.to(device)
sequence = sequence.to(device)
target = target.to(device)
outputs = model(minip, sequence)
pred = outputs[0].argmax(dim=1)
# Compute Dice score
for c in range(1, 3): # Classes 1 and 2 (BV and MAT)
pred_c = (pred == c).float()
target_c = (target == c).float()
intersection = (pred_c * target_c).sum()
union = pred_c.sum() + target_c.sum()
if union > 0:
dice = 2 * intersection / union
val_dice += dice.item()
val_samples += 1
avg_val_dice = val_dice / val_samples if val_samples > 0 else 0
print(f'Epoch {epoch}, Train Loss: {train_loss/len(train_loader):.4f}, '
f'Val Dice: {avg_val_dice:.4f}')
# Save best model
if avg_val_dice > best_val_dice:
best_val_dice = avg_val_dice
torch.save(model.state_dict(), 'best_dsanet_model.pth')
print(f'Saved best model with Dice: {best_val_dice:.4f}')
# Data preprocessing and loading
import cv2
from torch.utils.data import Dataset, DataLoader
import os
from skimage import io
import random
class DSCADataset(Dataset):
"""Dataset class for DSCA DSA sequences"""
def __init__(self, data_dir, split='train', transform=None, num_frames=8):
self.data_dir = data_dir
self.split = split
self.transform = transform
self.num_frames = num_frames
# Load dataset information
self.sequences = []
self.load_dataset_info()
def load_dataset_info(self):
"""Load sequence paths and annotations"""
# This is a placeholder - in practice, you would load from the actual DSCA dataset
split_file = os.path.join(self.data_dir, f'{self.split}.txt')
with open(split_file, 'r') as f:
for line in f:
seq_path, minip_path, mask_path = line.strip().split(',')
self.sequences.append({
'seq_path': seq_path,
'minip_path': minip_path,
'mask_path': mask_path
})
def load_sequence(self, seq_path):
"""Load and resample DSA sequence to fixed number of frames"""
# Load all frames in the sequence
frames = []
frame_files = sorted(os.listdir(seq_path))
for frame_file in frame_files:
frame = cv2.imread(os.path.join(seq_path, frame_file), cv2.IMREAD_GRAYSCALE)
frames.append(frame)
# Resample to fixed number of frames
if len(frames) > self.num_frames:
# Uniformly sample frames
indices = np.linspace(0, len(frames)-1, self.num_frames, dtype=int)
frames = [frames[i] for i in indices]
elif len(frames) < self.num_frames:
# Repeat frames if necessary
while len(frames) < self.num_frames:
frames.append(frames[-1])
return np.stack(frames)
def __len__(self):
return len(self.sequences)
def __getitem__(self, idx):
seq_info = self.sequences[idx]
# Load sequence
sequence = self.load_sequence(seq_info['seq_path'])
# Load MinIP image
minip = cv2.imread(seq_info['minip_path'], cv2.IMREAD_GRAYSCALE)
# Load mask (0: background, 1: BV, 2: MAT)
mask = cv2.imread(seq_info['mask_path'], cv2.IMREAD_GRAYSCALE)
# Apply augmentations if in training mode
if self.split == 'train' and self.transform:
# Random crop
h, w = minip.shape
crop_size = 512
if h > crop_size and w > crop_size:
y = random.randint(0, h - crop_size)
x = random.randint(0, w - crop_size)
minip = minip[y:y+crop_size, x:x+crop_size]
mask = mask[y:y+crop_size, x:x+crop_size]
sequence = sequence[:, y:y+crop_size, x:x+crop_size]
# Random horizontal flip
if random.random() > 0.5:
minip = np.fliplr(minip)
mask = np.fliplr(mask)
sequence = np.flip(sequence, axis=2)
# Random rotation
if random.random() > 0.5:
angle = random.uniform(-20, 20)
h, w = minip.shape
M = cv2.getRotationMatrix2D((w/2, h/2), angle, 1)
minip = cv2.warpAffine(minip, M, (w, h))
mask = cv2.warpAffine(mask, M, (w, h), flags=cv2.INTER_NEAREST)
for i in range(len(sequence)):
sequence[i] = cv2.warpAffine(sequence[i], M, (w, h))
# Normalize to [0, 1]
minip = minip.astype(np.float32) / 255.0
sequence = sequence.astype(np.float32) / 255.0
# Convert to tensors
minip = torch.from_numpy(minip).unsqueeze(0) # Add channel dimension
sequence = torch.from_numpy(sequence).unsqueeze(1) # Add channel dimension
mask = torch.from_numpy(mask).long()
return minip, sequence, mask
def create_data_loaders(data_dir, batch_size=2, num_workers=4):
"""Create train and validation data loaders"""
train_dataset = DSCADataset(data_dir, split='train', transform=True)
val_dataset = DSCADataset(data_dir, split='val', transform=False)
train_loader = DataLoader(
train_dataset,
batch_size=batch_size,
shuffle=True,
num_workers=num_workers,
pin_memory=True
)
val_loader = DataLoader(
val_dataset,
batch_size=batch_size,
shuffle=False,
num_workers=num_workers,
pin_memory=True
)
return train_loader, val_loader
# Inference function
def inference(model, minip_path, sequence_dir, device='cuda'):
"""Run inference on a single case"""
model.eval()
# Load and preprocess MinIP
minip = cv2.imread(minip_path, cv2.IMREAD_GRAYSCALE)
minip = minip.astype(np.float32) / 255.0
minip_tensor = torch.from_numpy(minip).unsqueeze(0).unsqueeze(0).to(device)
# Load and preprocess sequence
frames = []
frame_files = sorted(os.listdir(sequence_dir))
# Resample to 8 frames
indices = np.linspace(0, len(frame_files)-1, 8, dtype=int)
for i in indices:
frame = cv2.imread(os.path.join(sequence_dir, frame_files[i]), cv2.IMREAD_GRAYSCALE)
frame = frame.astype(np.float32) / 255.0
frames.append(frame)
sequence = np.stack(frames)
sequence_tensor = torch.from_numpy(sequence).unsqueeze(0).unsqueeze(2).to(device)
# Run inference
with torch.no_grad():
outputs = model(minip_tensor, sequence_tensor)
prediction = outputs[0].argmax(dim=1).squeeze().cpu().numpy()
return prediction
# Evaluation metrics
def compute_metrics(pred, target, num_classes=3):
"""Compute segmentation metrics"""
metrics = {}
for c in range(num_classes):
pred_c = (pred == c)
target_c = (target == c)
tp = np.sum(pred_c & target_c)
fp = np.sum(pred_c & ~target_c)
fn = np.sum(~pred_c & target_c)
# Dice
dice = 2 * tp / (2 * tp + fp + fn) if (2 * tp + fp + fn) > 0 else 0
# Jaccard
jaccard = tp / (tp + fp + fn) if (tp + fp + fn) > 0 else 0
# Sensitivity
sensitivity = tp / (tp + fn) if (tp + fn) > 0 else 0
# Precision
precision = tp / (tp + fp) if (tp + fp) > 0 else 0
class_name = ['Background', 'BV', 'MAT'][c]
metrics[f'{class_name}_Dice'] = dice
metrics[f'{class_name}_JAC'] = jaccard
metrics[f'{class_name}_SEN'] = sensitivity
metrics[f'{class_name}_PRE'] = precision
return metrics
# Main training script
def main():
# Configuration
data_dir = '/path/to/DSCA/dataset'
batch_size = 2
num_epochs = 500
device = 'cuda' if torch.cuda.is_available() else 'cpu'
# Create model
model = DSANet(num_classes=3)
print(f"Model created with {sum(p.numel() for p in model.parameters())} parameters")
# Create data loaders
train_loader, val_loader = create_data_loaders(data_dir, batch_size)
# Train model
train_dsanet(model, train_loader, val_loader, num_epochs, device)
# Load best model for testing
model.load_state_dict(torch.load('best_dsanet_model.pth'))
model.to(device)
# Example inference
pred = inference(model, 'test_minip.png', 'test_sequence_dir/', device)
print(f"Prediction shape: {pred.shape}")
# Example usage
if __name__ == "__main__":
# For demonstration, create a small example
model = DSANet(num_classes=3)
# Example input
batch_size = 2
minip = torch.randn(batch_size, 1, 512, 512)
sequence = torch.randn(batch_size, 8, 1, 512, 512)
# Forward pass
outputs = model(minip, sequence)
main_out, out2, out3 = outputs
print(f"Main output shape: {main_out.shape}")
print(f"Deep supervision output 2 shape: {out2.shape}")
print(f"Deep supervision output 3 shape: {out3.shape}")
# Compute loss
target = torch.randint(0, 3, (batch_size, 512, 512))
criterion = CombinedLoss(num_classes=3)
loss = criterion(outputs, target)
print(f"Loss: {loss.item():.4f}")
# Run full training (comment out for quick test)
# main()