Cancer remains one of the most formidable challenges in modern medicine. While immunotherapy has revolutionized treatment for some, many tumors—known as “cold tumors”—remain unresponsive. These tumors lack the immune cell infiltration necessary for therapies like checkpoint inhibitors to work. The key to unlocking their potential? Immunogenic Cell Death (ICD).
ICD is a powerful form of regulated cell death that doesn’t just kill cancer cells—it turns them into a beacon for the immune system. When cancer cells die immunogenically, they release signals called Damage-Associated Molecular Patterns (DAMPs) such as calreticulin (CRT), ATP, and HMGB1. These molecules act like alarm bells, recruiting dendritic cells, promoting antigen presentation, and ultimately turning a cold tumor into a “hot,” immune-responsive one.
But identifying drugs that induce ICD has traditionally been slow, expensive, and limited. Enter artificial intelligence (AI)—a game-changer that’s not only accelerating discovery but doing so with unprecedented accuracy.
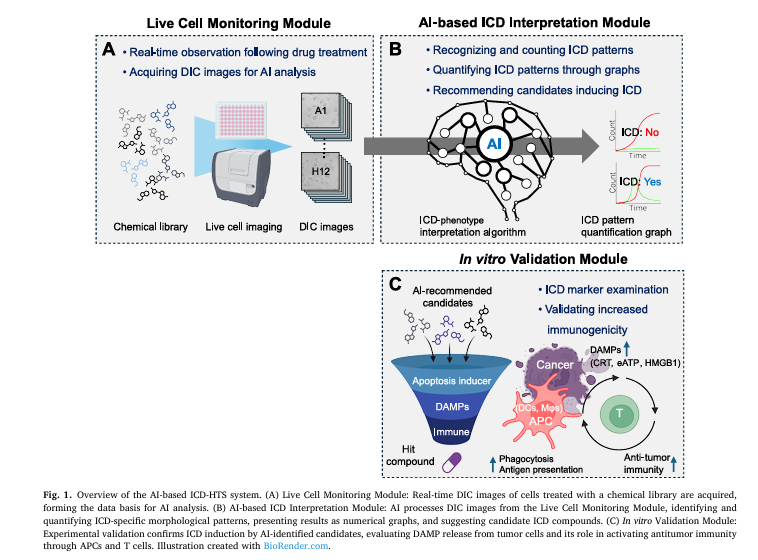
In a groundbreaking 2025 study published in Computers in Biology and Medicine, researchers from Inha University unveiled an AI-powered high-throughput screening (HTS) system that can identify ICD inducers using only real-time optical images—no fluorescent labels required. This innovation is poised to redefine how we discover next-generation cancer immunotherapies.
Let’s dive into the 7 revolutionary aspects of this new system—and why outdated screening methods are falling behind.
1. The Problem: Traditional ICD Screening is Slow, Costly, and Incomplete
Before we explore the solution, it’s crucial to understand what’s wrong with current approaches.
Most high-throughput screening for ICD relies on fluorescence-based assays or luminescence measurements to detect DAMPs like ATP or HMGB1. While useful, these methods have critical flaws:
- 🔬 They’re endpoint assays – You only get a snapshot at a single time point, missing the dynamic progression of cell death.
- 💸 They’re expensive – Fluorescent tags, antibodies, and reagents drive up costs.
- 🧫 They’re low-resolution – They fail to capture subtle morphological changes like cell swelling or membrane rupture in real time.
- 🕒 They’re labor-intensive – Manual image analysis is slow and prone to human error.
As the study notes: “Interpreting these complex image data remains time-consuming and labor-intensive, and traditional methods often prove to be inefficient for quantification.”
This bottleneck has severely limited the pace of ICD drug discovery.
2. The Solution: An AI-Powered HTS System for Real-Time ICD Detection
The new system developed by Kim et al. is a three-module AI pipeline designed to overcome these limitations:
| MODULE | FUNCTION | INNOVATION |
|---|---|---|
| 1. Live Cell Monitoring | Captures real-time DIC images of cells treated with drug candidates | Label-free, continuous imaging |
| 2. AI-Based ICD Interpretation | Uses deep learning to detect ICD-specific morphological changes | No fluorescence needed |
| 3. In Vitro Validation | Confirms DAMP release and immune activation | Ensures biological relevance |
This integrated workflow enables automated, scalable, and accurate screening of ICD inducers—without relying on expensive fluorescent markers.
3. How It Works: From DIC Images to ICD Prediction
The system uses Differential Interference Contrast (DIC) microscopy, a label-free imaging technique that visualizes live cells in high contrast based on optical density.
But DIC images alone are challenging for AI to interpret. So the researchers used a transfer learning strategy:
- Pretraining on Fluorescent Data: The AI model was first trained on multichannel images (DIC + GFP + RFP) from cells expressing HMGB1-tGFP and stained with SYTOX Orange.
- Fine-Tuning on DIC Only: After learning the morphological signatures of ICD, the model was fine-tuned using only DIC images.
This allowed the AI to “see” ICD patterns in unlabeled images—like a radiologist trained on annotated X-rays, now reading new scans blind.
4. Key Morphological Features of ICD Identified by AI
The AI was trained to detect two critical stages of ICD:
- Swelling Cells (SC): Cells that have started to swell due to membrane stress but still have intact membranes.
- Dead Cells (DC): Cells with ruptured membranes, releasing DAMPs.
These stages are visually distinct in DIC:
- SCs appear larger, phase-bright, and rounded.
- DCs show loss of structure and membrane fragmentation.
The AI quantifies the ratio of SC to DC over time, creating kinetic profiles that reveal whether a compound induces ICD.
🔍 Why this matters: A true ICD inducer shows a clear peak in SCs before DCs—indicating a controlled, immunogenic process. Apoptotic agents, in contrast, often skip swelling and go straight to death.
5. Model-Assisted Labeling (MAL): 4x Faster Annotation
One of the biggest bottlenecks in AI training is data labeling. Manually annotating thousands of cells is slow and error-prone.
The team introduced Model-Assisted Labeling (MAL), an iterative process:
- A small set of cells is manually labeled.
- The AI uses this to make initial predictions on new images.
- Experts correct only the uncertain or incorrect predictions.
- The corrected data is fed back to retrain the model.
Results?
- Manual labeling: 4.3–4.7 seconds per cell
- MAL-assisted labeling: 1.04 seconds per cell — a 76% reduction
- More cells labeled: 1,490 vs. ~1,100–1,300
This not only speeds up training but improves consistency and recall.
6. Performance: AI Outperforms Human Experts
The final AI model was evaluated using standard metrics:
| MODEL | SC F1-SCORE | SC MAP50 | DC F1-SCORE | DC MAP50 |
|---|---|---|---|---|
| DIC-Only | 0.444 | 0.255 | 0.754 | 0.634 |
| Fine-Tuned | 0.470 | 0.287 | 0.758 | 0.644 |
| Final (Ensemble + Post-Processing) | 0.509 | 0.349 | 0.794 | 0.721 |
The 6.5% improvement in SC F1 score is significant—especially since swelling cells are the earliest sign of ICD.
To reduce false positives (e.g., debris misclassified as SC), the team added an ML-based post-processing step using XGBoost to filter predictions based on bounding box dimensions.
The classifier uses a log-odds decision function:
$$y^{i} = \begin{cases} SC,\;\text{debris}, & \text{if}\; \sigma\left(\sum_{k=1}^{K} \eta f_{k}(x^{i})\right) \geq \theta \\ \text{otherwise} \end{cases}$$Where:
- fk (xi) : output of the k -th decision tree
- η : learning rate
- σ(⋅) : sigmoid function
- θ : decision threshold (e.g., 0.5)
This step significantly improved precision without sacrificing recall.
7. Blind Test: AI Correctly Identifies 3 of 8 ICD Inducers
In a real-world validation, the AI was given 8 unknown compounds and asked to classify them as ICD inducers.
The AI analyzed time-series DIC images and generated SC/DC ratio graphs. It identified compounds B, D, G, and H as ICD candidates based on the presence of an SC peak before DC increase.
In vitro validation confirmed:
- Compounds B (Erastin), D (FIN56), and H (RSL3) induced ferroptosis—a known ICD pathway.
- Compound G (Raptinal) showed mixed activity: it induced apoptosis but also pyroptosis and DAMP release, confirming recent findings that it can trigger ICD.
| COMPOUND | AI PREDICTION | CELL DEATH TYPE | ICD CONFIRMED? |
|---|---|---|---|
| B (Erastin) | ✅ | Ferroptosis | ✅ |
| D (FIN56) | ✅ | Ferroptosis | ✅ |
| H (RSL3) | ✅ | Ferroptosis | ✅ |
| G (Raptinal) | ✅ | Apoptosis + Pyroptosis | ✅ (partial) |
| Others (A, C, E, F) | ❌ | Apoptosis | ❌ |
This demonstrates the AI’s ability to detect complex, mixed cell death mechanisms—something human experts might miss.
Why This Matters: The Future of Cancer Drug Discovery
This AI system isn’t just faster—it’s smarter. It can:
- Detect subtle morphological differences invisible to the human eye.
- Predict optimal dosing by analyzing the timing of SC peaks.
- Identify dual-mechanism drugs like raptinal that induce both apoptosis and ICD.
- Reduce screening costs by eliminating fluorescent reagents.
As the authors state: “Our AI-based HTS system efficiently identified ICD candidates using only real-time optical images, thereby significantly reducing the time and resources required for screening.”
Limitations and the Road Ahead
Despite its success, the system has limitations:
- Trained primarily on MDA-MB-231 cells and ferroptosis inducers.
- May not generalize to other cell lines or ICD pathways (e.g., pyroptosis, necroptosis).
Future work should:
- Expand training data to include diverse cell types and death pathways.
- Enable multi-class classification (e.g., apoptosis vs. ferroptosis vs. pyroptosis).
- Integrate single-cell tracking to study heterogeneity in drug response.
Conclusion: A New Era in Immunotherapy Screening
The fusion of AI and real-time imaging is transforming how we discover cancer drugs. This study proves that label-free, high-throughput ICD screening is not only possible—it’s superior to traditional methods.
By automating the detection of immunogenic cell death, this system accelerates the development of therapies that can turn cold tumors hot, making them vulnerable to the immune system.
For researchers, pharmaceutical companies, and patients alike, this is more than a technical advance—it’s a hope multiplier.
If you’re Interested in Melanoma Detection with AI, you may also find this article helpful: 7 Revolutionary Breakthroughs in Melanoma Diagnosis: The Quantum AI Edge That’s Changing Everything
Call to Action: Join the AI Revolution in Cancer Research
Are you working on cancer immunotherapy or drug discovery? Leverage this breakthrough today.
👉 Access the open-source code and start building your own AI-powered screening pipeline:
https://github.com/MAI00024/2025-ICD
📊 Download the biological dataset to validate and extend the model:
https://gofile.me/7lgf2/p45mpeT6i
📩 Subscribe to our newsletter for updates on AI in biomedicine and exclusive access to upcoming tools.
📊 Read the Complete Paper: High-throughput screening system for immunogenic cell death inducers using artificial intelligence-based real-time image analysis
The future of cancer therapy isn’t just in the lab—it’s in the algorithm. Don’t get left behind.
Here is the end-to-end Python code that implements the proposed model.
#
# End-to-End Python Implementation of the AI-based ICD Screening System
# Based on the paper: "High-throughput screening system for immunogenic cell death
# inducers using artificial intelligence-based real-time image analysis"
#
# This script simulates the complete pipeline described in the paper, including:
# 1. Data Preparation & Preprocessing
# 2. Model-Assisted Labeling (MAL)
# 3. Fluorescence-based Pre-training (FPDF)
# 4. DIC-based Fine-tuning
# 5. ML-based Post-processing for debris filtering
# 6. Inference on new images
#
import numpy as np
import torch
import torchvision
from torchvision.models.detection import fasterrcnn_resnet50_fpn
from torchvision.transforms import functional as F
import xgboost as xgb
from sklearn.model_selection import train_test_split
import cv2 # OpenCV for image manipulation
import os
import random
# --- Configuration ---
# Note: The paper uses YOLOv9. As YOLOv9 is not a standard PyTorch library,
# we use Faster R-CNN as a stand-in to make this code runnable.
# The logic remains the same and can be adapted for a YOLOv9 implementation.
DEVICE = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
NUM_CLASSES = 3 # 0: background, 1: Swelling Cell (SC), 2: Dead Cell (DC)
MODEL_SAVE_PATH = "./models"
os.makedirs(MODEL_SAVE_PATH, exist_ok=True)
# --- Helper Functions for Data Simulation ---
def create_dummy_image(size=(1024, 1024), num_cells=50):
"""Creates a blank image with some dummy 'cells' (circles)."""
image = np.full((size[0], size[1], 3), 240, dtype=np.uint8) # Light gray background like DIC
boxes = []
labels = []
for _ in range(num_cells):
label = random.randint(1, NUM_CLASSES - 1) # 1 for SC, 2 for DC
cx, cy = random.randint(50, size[0] - 50), random.randint(50, size[1] - 50)
radius = random.randint(15, 30)
color = (180, 180, 180) if label == 1 else (100, 100, 100) # SC is lighter, DC is darker
cv2.circle(image, (cx, cy), radius, color, -1)
# Create bounding box
x_min, y_min = cx - radius, cy - radius
x_max, y_max = cx + radius, cy + radius
boxes.append([x_min, y_min, x_max, y_max])
labels.append(label)
return image, boxes, labels
def create_dummy_fluorescence_image(size=(1024, 1024), num_cells=50):
"""
Creates a dummy multi-channel image simulating merged DIC, GFP, and RFP.
- SC: Green channel (GFP) is active.
- DC: Red channel (RFP) is active.
"""
# Start with a DIC-like base image (grayscale)
dic_channel = np.full((size[0], size[1]), 240, dtype=np.uint8)
gfp_channel = np.zeros((size[0], size[1]), dtype=np.uint8)
rfp_channel = np.zeros((size[0], size[1]), dtype=np.uint8)
boxes = []
labels = []
for _ in range(num_cells):
label = random.randint(1, NUM_CLASSES - 1)
cx, cy = random.randint(50, size[0] - 50), random.randint(50, size[1] - 50)
radius = random.randint(15, 30)
# Draw on DIC channel
cv2.circle(dic_channel, (cx, cy), radius, 150, -1)
# Draw on fluorescence channels
if label == 1: # Swelling Cell (SC) -> GFP
cv2.circle(gfp_channel, (cx, cy), radius, 255, -1)
else: # Dead Cell (DC) -> RFP
cv2.circle(rfp_channel, (cx, cy), radius, 255, -1)
x_min, y_min = cx - radius, cy - radius
x_max, y_max = cx + radius, cy + radius
boxes.append([x_min, y_min, x_max, y_max])
labels.append(label)
# Merge channels into an RGB-like image
merged_image = cv2.merge([rfp_channel, gfp_channel, dic_channel])
return merged_image, boxes, labels
class ICDDataset(torch.utils.data.Dataset):
"""Custom PyTorch Dataset for ICD data."""
def __init__(self, images, targets):
self.images = images
self.targets = targets
def __getitem__(self, idx):
image = self.images[idx]
target = self.targets[idx]
# Convert to tensor
image_tensor = F.to_tensor(image)
# Format targets for Faster R-CNN
target_dict = {
'boxes': torch.as_tensor(target['boxes'], dtype=torch.float32),
'labels': torch.as_tensor(target['labels'], dtype=torch.int64),
}
return image_tensor, target_dict
def __len__(self):
return len(self.images)
def collate_fn(batch):
"""Collate function for the DataLoader."""
return tuple(zip(*batch))
class ICDScreeningSystem:
"""
Main class to encapsulate the entire AI-based screening pipeline.
"""
def __init__(self):
self.detection_model = None
self.postprocessing_classifier = None
print("ICD Screening System initialized.")
def _get_model(self, num_classes):
"""Loads the object detection model."""
# The paper uses YOLOv9. We use Faster R-CNN as a stand-in.
# To use YOLOv9, you would load its specific architecture here.
model = fasterrcnn_resnet50_fpn(weights="FasterRCNN_ResNet50_FPN_Weights.DEFAULT")
in_features = model.roi_heads.box_predictor.cls_score.in_features
model.roi_heads.box_predictor = torchvision.models.detection.faster_rcnn.FastRCNNPredictor(in_features, num_classes)
return model.to(DEVICE)
def train_detection_model(self, dataset, num_epochs=10, model_name="model.pth"):
"""
Handles the training loop for the object detection model.
This is used for both pre-training and fine-tuning.
"""
print(f"--- Starting training for {model_name} ---")
self.detection_model = self._get_model(NUM_CLASSES)
data_loader = torch.utils.data.DataLoader(dataset, batch_size=2, shuffle=True, collate_fn=collate_fn)
params = [p for p in self.detection_model.parameters() if p.requires_grad]
optimizer = torch.optim.SGD(params, lr=0.005, momentum=0.9, weight_decay=0.0005)
for epoch in range(num_epochs):
self.detection_model.train()
epoch_loss = 0
for images, targets in data_loader:
images = list(image.to(DEVICE) for image in images)
targets = [{k: v.to(DEVICE) for k, v in t.items()} for t in targets]
loss_dict = self.detection_model(images, targets)
losses = sum(loss for loss in loss_dict.values())
optimizer.zero_grad()
losses.backward()
optimizer.step()
epoch_loss += losses.item()
print(f"Epoch {epoch+1}/{num_epochs}, Loss: {epoch_loss/len(data_loader):.4f}")
torch.save(self.detection_model.state_dict(), os.path.join(MODEL_SAVE_PATH, model_name))
print(f"--- Model saved to {os.path.join(MODEL_SAVE_PATH, model_name)} ---")
def model_assisted_labeling(self, unlabeled_images, initial_model_path=None):
"""
Simulates the Model-Assisted Labeling (MAL) process.
In a real scenario, this would involve a human-in-the-loop to verify predictions.
"""
print("\n--- Starting Model-Assisted Labeling (MAL) ---")
if initial_model_path:
model = self._get_model(NUM_CLASSES)
model.load_state_dict(torch.load(initial_model_path))
model.eval()
print("Loaded initial model for pseudo-labeling.")
else:
print("No initial model provided. MAL would require manual labeling first.")
return []
pseudo_labeled_data = []
for i, image in enumerate(unlabeled_images):
image_tensor = F.to_tensor(image).unsqueeze(0).to(DEVICE)
with torch.no_grad():
predictions = model(image_tensor)[0]
# Here, an expert would review `predictions` (boxes, labels, scores)
# and correct them. We simulate this by accepting high-confidence predictions.
boxes = predictions['boxes'][predictions['scores'] > 0.8].cpu().numpy().tolist()
labels = predictions['labels'][predictions['scores'] > 0.8].cpu().numpy().tolist()
print(f"Image {i}: Generated {len(boxes)} pseudo-labels for expert review.")
# This corrected data would be added to the training set.
if boxes:
pseudo_labeled_data.append({'image': image, 'target': {'boxes': boxes, 'labels': labels}})
print("--- MAL process complete. The pseudo-labeled data can now be used for re-training. ---")
return pseudo_labeled_data
def train_postprocessing_classifier(self, labeled_dataset, debris_dataset):
"""
Trains an XGBoost classifier to distinguish between Swelling Cells (SC) and debris.
"""
print("\n--- Training Post-processing Classifier (Debris Filtering) ---")
# Extract features (width, height) from SC bounding boxes
sc_features = []
for _, target in labeled_dataset:
for i, label in enumerate(target['labels']):
if label == 1: # Label 1 is SC
box = target['boxes'][i]
width = box[2] - box[0]
height = box[3] - box[1]
sc_features.append([width, height])
# Simulate debris features (typically smaller and more erratic)
debris_features = []
for _, target in debris_dataset:
for box in target['boxes']:
width = box[2] - box[0]
height = box[3] - box[1]
debris_features.append([width, height])
print(f"Found {len(sc_features)} SC instances and {len(debris_features)} debris instances.")
# Create dataset for XGBoost
X = np.array(sc_features + debris_features)
y = np.array([1] * len(sc_features) + [0] * len(debris_features)) # 1 for SC, 0 for Debris
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
self.postprocessing_classifier = xgb.XGBClassifier(objective='binary:logistic', use_label_encoder=False, eval_metric='logloss')
self.postprocessing_classifier.fit(X_train, y_train)
accuracy = self.postprocessing_classifier.score(X_test, y_test)
print(f"Post-processing classifier trained with accuracy: {accuracy:.4f}")
def predict(self, dic_image, model_path, confidence_threshold=0.7):
"""
Performs inference on a single DIC image, including post-processing.
"""
print("\n--- Running Inference on a new DIC image ---")
model = self._get_model(NUM_CLASSES)
model.load_state_dict(torch.load(model_path))
model.to(DEVICE)
model.eval()
image_tensor = F.to_tensor(dic_image).unsqueeze(0).to(DEVICE)
with torch.no_grad():
predictions = model(image_tensor)[0]
# Initial predictions
boxes = predictions['boxes'][predictions['scores'] > confidence_threshold]
labels = predictions['labels'][predictions['scores'] > confidence_threshold]
print(f"Initial detection: Found {len(boxes)} objects.")
# Apply post-processing to filter SC predictions
final_boxes = []
final_labels = []
if self.postprocessing_classifier is None:
print("Warning: Post-processing classifier not trained. Skipping debris filtering.")
return boxes.cpu().numpy(), labels.cpu().numpy()
for box, label in zip(boxes, labels):
if label == 1: # If it's a Swelling Cell, verify with classifier
width = box[2] - box[0]
height = box[3] - box[1]
feature = np.array([[width.cpu(), height.cpu()]])
# Predict if it's SC (1) or Debris (0)
if self.postprocessing_classifier.predict(feature)[0] == 1:
final_boxes.append(box.cpu().numpy())
final_labels.append(label.cpu().numpy())
else: # If it's a Dead Cell (label 2), keep it
final_boxes.append(box.cpu().numpy())
final_labels.append(label.cpu().numpy())
num_filtered = len(boxes) - len(final_boxes)
print(f"Post-processing: Filtered out {num_filtered} potential debris objects.")
print(f"Final detection: {len(final_labels)} objects ({final_labels.count(1)} SC, {final_labels.count(2)} DC).")
return final_boxes, final_labels
# --- Main Execution: Simulating the End-to-End Pipeline ---
if __name__ == '__main__':
# 1. DATA PREPARATION (SIMULATED)
print("Step 1: Simulating Data Preparation\n")
# Create a small manually labeled dataset to start
manual_images = []
manual_targets = []
for _ in range(5): # 5 initial images
img, b, l = create_dummy_image()
manual_images.append(img)
manual_targets.append({'boxes': b, 'labels': l})
# Create a larger pool of unlabeled data for MAL
unlabeled_dic_pool = [create_dummy_image()[0] for _ in range(20)]
# Create fluorescence data for pre-training
fluorescence_images = []
fluorescence_targets = []
for _ in range(10): # 10 fluorescence images
img, b, l = create_dummy_fluorescence_image()
fluorescence_images.append(img)
fluorescence_targets.append({'boxes': b, 'labels': l})
# Create simulated debris data for post-processing classifier
debris_images, debris_targets = [], []
for _ in range(5):
# Debris are just small, random boxes
img, _, _ = create_dummy_image(num_cells=0)
debris_b = [[random.randint(100, 900), random.randint(100, 900), random.randint(100, 900)+random.randint(5,10), random.randint(100, 900)+random.randint(5,10)] for _ in range(20)]
debris_images.append(img)
debris_targets.append({'boxes': debris_b, 'labels': [1]*len(debris_b)}) # Falsely labeled as SC
# 2. FPDF PART 1: PRE-TRAINING ON FLUORESCENCE DATA
print("\nStep 2: Pre-training on Fluorescence Data (FPDF Stage 1)")
system = ICDScreeningSystem()
fluorescence_dataset = ICDDataset(fluorescence_images, fluorescence_targets)
system.train_detection_model(fluorescence_dataset, num_epochs=5, model_name="pretrained_fluorescence_model.pth")
# 3. MODEL-ASSISTED LABELING (MAL)
# Use the pre-trained model to help label more DIC images
print("\nStep 3: Model-Assisted Labeling (MAL)")
pseudo_labeled_data = system.model_assisted_labeling(
unlabeled_dic_pool,
initial_model_path=os.path.join(MODEL_SAVE_PATH, "pretrained_fluorescence_model.pth")
)
# Combine manual and pseudo-labeled data for fine-tuning
all_dic_images = manual_images + [item['image'] for item in pseudo_labeled_data]
all_dic_targets = manual_targets + [item['target'] for item in pseudo_labeled_data]
# 4. FPDF PART 2: FINE-TUNING ON DIC DATA
print("\nStep 4: Fine-tuning on DIC Data (FPDF Stage 2)")
dic_dataset = ICDDataset(all_dic_images, all_dic_targets)
system.train_detection_model(dic_dataset, num_epochs=5, model_name="finetuned_dic_model.pth")
# 5. TRAIN POST-PROCESSING CLASSIFIER
print("\nStep 5: Training Post-processing Debris Filter")
# We use the full DIC dataset as positive examples and simulated debris as negative
system.train_postprocessing_classifier(dic_dataset, ICDDataset(debris_images, debris_targets))
# 6. INFERENCE
print("\nStep 6: Running Inference on a new test image")
test_image, true_boxes, true_labels = create_dummy_image(num_cells=60)
final_boxes, final_labels = system.predict(
test_image,
model_path=os.path.join(MODEL_SAVE_PATH, "finetuned_dic_model.pth")
)
# Visualize the result
output_image = test_image.copy()
for box, label in zip(final_boxes, final_labels):
x1, y1, x2, y2 = map(int, box)
color = (0, 255, 0) if label == 1 else (0, 0, 255) # Green for SC, Red for DC
label_text = "SC" if label == 1 else "DC"
cv2.rectangle(output_image, (x1, y1), (x2, y2), color, 2)
cv2.putText(output_image, label_text, (x1, y1 - 10), cv2.FONT_HERSHEY_SIMPLEX, 0.7, color, 2)
cv2.imwrite("output_detection_result.png", output_image)
print("\n--- End of Pipeline ---")
print("Inference result saved to 'output_detection_result.png'")