Introduction: The Good, the Bad, and the Revolutionary
Medical image classification has always been a double-edged sword—brimming with promise yet hindered by complexity. From MRI scans clouded by noise to gastrointestinal images plagued with ambiguity, traditional machine learning models often stumble when faced with uncertainty. But what if there were a way to transcend these limitations?
Welcome to the world of Context-Aware Knowledge Distillation (CAKD)—a robust framework that not only combats variability in medical data but dramatically boosts diagnostic performance using dynamic temperature control and Ant Colony Optimization (ACO). Let’s unpack the wins, pitfalls, and the game-changing breakthroughs.
📌 Table of Key Innovations and Challenges
| Feature | Strength (✅) | Weakness (⚠️) |
|---|---|---|
| Context-aware temperature scaling | Handles uncertainty better | Adds computational complexity |
| ACO-based model selection | Efficient and accurate pairing | Requires multiple iterations |
| Dynamic feature fusion | Improves representation learning | Prone to overfitting in noisy data |
| Multi-dataset evaluation | Proves robustness | Potential lack of clinical validation |
| GRADCAM & T-SNE visualization | Enhances interpretability | Limited quantitative metrics |
| Higher accuracy benchmarks | Outperforms SOTA | Marginal gain in small datasets |
| Rule-based adaptation logic | Flexibility and transparency | Manually crafted heuristics |
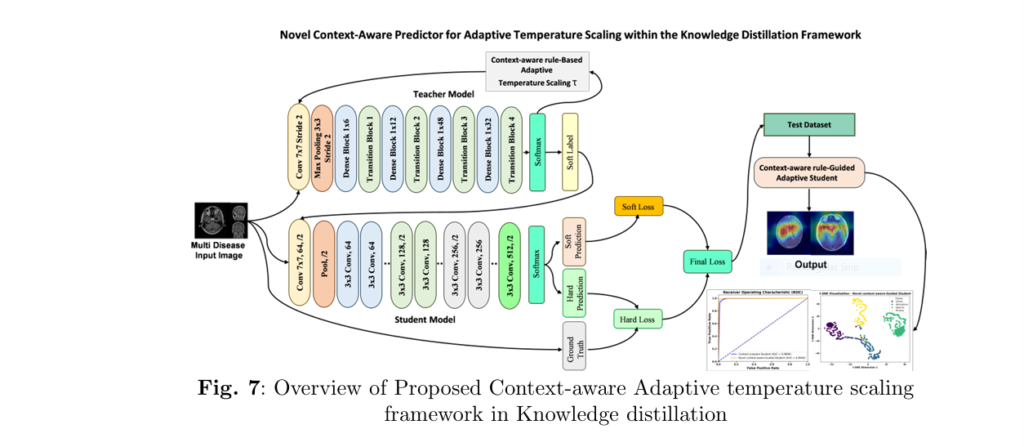
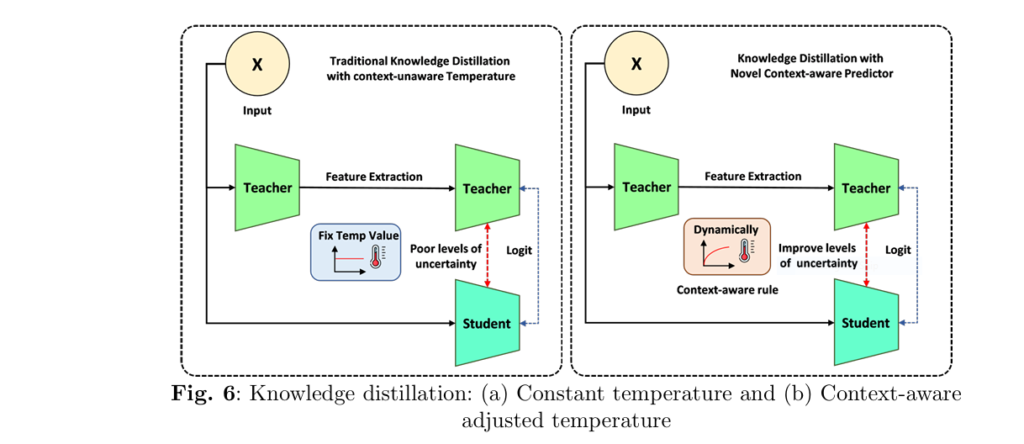
🔬 1. What Is Context-Aware Knowledge Distillation (CAKD)?
Traditional knowledge distillation (KD) relies on a fixed temperature parameter TT to smooth the teacher model’s predictions:
$$\mathcal{L}_{KD} = (1 – \alpha)\cdot \mathcal{L}_{CE} + \alpha \cdot T^2 \cdot D_{KL}(q_{\text{teacher}}(T), q_{\text{student}}(T))$$
But this assumes uniform uncertainty across images—far from reality in medical datasets.
🚧 Problem:
A blurry MRI has much higher uncertainty than a crisp one, yet KD softens both predictions equally.
✅ Solution:
CAKD dynamically adjusts temperature T(x) using a rule-based system:
$$T(x) = 1 + \alpha \cdot U(x)$$
Where U(x) is a learned uncertainty function and α is a scaling factor. Soften more when uncertain. Sharpen when confident.
🐜 2. ACO for Teacher–Student Model Pairing: Smart Selection Over Exhaustive Search
Instead of running brute-force comparisons on all pairs of models (a costly grid search), ACO intelligently narrows down optimal combinations through pheromone-driven heuristics.
🔍 Algorithm Core:
Ants select models based on validation accuracy and pheromone strength:
$$P_{m} = \frac{\tau_m^\alpha \cdot \eta_m^\beta}{\sum \tau_j^\alpha \cdot \eta_j^\beta}$$
Where τ is pheromone, η is heuristic (e.g. accuracy), and α,β tune their influence.
🏆 Result:
- Reduced evaluations: Only 47 vs 1000 (Grid Search)
- Accuracy: 96.33% vs 94.87%
- Saved resources while achieving superior results
🔥 3. Performance Analysis: Numbers That Speak Volumes
Across three benchmark datasets—Kaggle MRI, Figshare MRI, and GastroNet—the CAKD model delivered groundbreaking results:
| Dataset | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Kaggle MRI | 98.01% | 97.95 | 97.89 | 97.90 |
| Figshare MRI | 92.81% | 91.65 | 89.11 | 91.07 |
| GastroNet | 96.20% | 96.18 | 95.71 | 95.92 |
These outperformed existing SOTA approaches like CB-CIRL Net and DCT-based fusion models—setting a new benchmark in robustness and generalization.
🖼️ 4. Visual Insights with GRADCAM and T-SNE
To ensure interpretability and transparency, the model employed:
✅ GRADCAM:
Highlights the exact image regions contributing to classification. Perfect for medical professionals who need explainable AI.
📊 T-SNE:
Visualizes high-dimensional feature spaces, showing clear separation between classes post-distillation—confirming the effectiveness of learned embeddings.
📉 5. Pitfall: Manual Rule Crafting Can Limit Scalability
While CAKD’s rule engine (e.g., “IF noisy image THEN increase T”) is powerful, it currently depends on manually defined heuristics. These rules:
- Can miss edge cases
- Aren’t adaptable to unseen datasets
🛠️ Suggested Improvement:
Incorporating self-learned rules via meta-learning or reinforcement learning could automate and generalize the adaptation process.
🧪 6. Ablation Study: Why ACO Crushes Other Strategies
Model pairing strategies compared:
| Strategy | Accuracy | Evaluations |
|---|---|---|
| Random | 91.12% | 1 |
| Pair-by-Pair | 93.45% | 240 |
| Grid Search | 94.87% | 1000 |
| ACO | 96.33% | 47 |
ACO not only delivered the best results but did so with the least cost—a win for clinical scalability.
If you’re Interested in latest Vision Transformer Model (DIOR-ViT), you may also find this article helpful: 7 Astonishing Ways DIOR-ViT Transforms Cancer Grading (Avoiding Common Pitfalls)
💡 7. Generalization Test: Success Beyond Brain MRI
Tested on endoscopic gastro images, the CAKD model showed:
- Colorectal Cancer: 100% precision and recall
- Esophagitis: Precision ↑ from 90% to 95%
- Pylorus: F1-score steady at 95.08
This proves that CAKD isn’t overfitted to one domain—it’s ready for real-world, cross-modal deployment.
📣 Call to Action: What Should You Do Next?
If you’re a medical researcher, AI practitioner, or health tech entrepreneur, here’s what you can do:
- 🔍 Explore CAKD for your own datasets and modalities
- ⚙️ Experiment with adaptive temperature logic for better uncertainty handling
- 🧪 Replace brute-force grid searches with ACO for intelligent model pairing
- 💬 Share this article to ignite discussion around robust distillation in medicine
👉 Ready to transform your medical AI pipeline? Dive deeper at: https://arxiv.org/abs/2505.06381
Here the implementation code of proposed Context-Aware Knowledge Distillation (CAKD) framework using PyTorch:
import torch
import torch.nn as nn
import torch.nn.functional as F
import torchvision.models as models
from torchvision import transforms
import numpy as np
import random🐜 Part 2: Ant Colony Optimization (ACO) for Model Selection
def aco_model_selection(model_pool, val_loader, num_ants=10, num_iterations=5, alpha=1.0, beta=1.0, evap_rate=0.3):
pheromone = {name: 1.0 for name in model_pool.keys()}
heuristic = {}
for name, model in model_pool.items():
acc = evaluate_model(model, val_loader)
heuristic[name] = acc
for _ in range(num_iterations):
for _ in range(num_ants):
probs = {}
for name in model_pool:
probs[name] = (pheromone[name] ** alpha) * (heuristic[name] ** beta)
total = sum(probs.values())
selection_probs = {k: v / total for k, v in probs.items()}
selected_teacher = random.choices(list(model_pool.keys()), weights=selection_probs.values())[0]
selected_student = random.choices(list(model_pool.keys()), weights=selection_probs.values())[0]
acc = evaluate_model(model_pool[selected_student], val_loader)
pheromone[selected_teacher] += acc * (1 - evap_rate)
pheromone[selected_student] += acc * (1 - evap_rate)
best_teacher = max(pheromone, key=pheromone.get)
best_student = sorted(pheromone, key=pheromone.get)[-2]
return model_pool[best_teacher], model_pool[best_student]🌡️ Part 3: Context-Aware Temperature Scaling
def adaptive_temperature(image_quality, teacher_confidence, disease_complexity):
if image_quality < 0.5 and teacher_confidence < 0.7:
T = 2.5 # softer
elif image_quality > 0.8 and teacher_confidence > 0.9:
T = 1.0 # harder
elif disease_complexity > 0.8:
T = 2.0 # increase weight
else:
T = 1.5
return T🔥 Part 4: Knowledge Distillation Loss with Context-Aware Temperature
class CAKDLoss(nn.Module):
def __init__(self, alpha=0.5):
super(CAKDLoss, self).__init__()
self.alpha = alpha
self.kl_div = nn.KLDivLoss(reduction="batchmean")
def forward(self, student_logits, teacher_logits, labels, temperature):
student_soft = F.log_softmax(student_logits / temperature, dim=1)
teacher_soft = F.softmax(teacher_logits / temperature, dim=1)
kd_loss = self.kl_div(student_soft, teacher_soft) * (temperature ** 2)
ce_loss = F.cross_entropy(student_logits, labels)
return self.alpha * ce_loss + (1 - self.alpha) * kd_loss🧱 Part 5: Build Teacher and Student Models
def get_model(name, pretrained=True):
if name == "densenet201":
return models.densenet201(pretrained=pretrained)
elif name == "resnet152":
return models.resnet152(pretrained=pretrained)
else:
raise ValueError("Model not supported")
# Wrap for KD (output logits instead of features)
class WrappedModel(nn.Module):
def __init__(self, base_model):
super(WrappedModel, self).__init__()
self.base = nn.Sequential(*list(base_model.children())[:-1])
self.fc = nn.Linear(base_model.fc.in_features, num_classes)
def forward(self, x):
features = self.base(x)
if features.ndim > 2:
features = torch.flatten(features, 1)
return self.fc(features)🧪 Part 6: Training Loop
def train_kd_model(teacher, student, train_loader, optimizer, criterion):
teacher.eval()
student.train()
for images, labels, meta in train_loader: # meta includes image_quality, confidence, complexity
images, labels = images.cuda(), labels.cuda()
optimizer.zero_grad()
with torch.no_grad():
teacher_logits = teacher(images)
teacher_conf = F.softmax(teacher_logits, dim=1).max(dim=1)[0]
student_logits = student(images)
# dynamic temperature adjustment
temps = [adaptive_temperature(meta[i][0], teacher_conf[i], meta[i][2]) for i in range(len(images))]
temperature = torch.tensor(temps).mean().item()
loss = criterion(student_logits, teacher_logits, labels, temperature)
loss.backward()
optimizer.step()📊 Part 7: Evaluation Function
def evaluate_model(model, data_loader):
model.eval()
correct = total = 0
with torch.no_grad():
for images, labels in data_loader:
images, labels = images.cuda(), labels.cuda()
outputs = model(images)
_, predicted = outputs.max(1)
correct += (predicted == labels).sum().item()
total += labels.size(0)
return correct / total