Detecting tiny vehicles in drone footage. Spotting distant pedestrians in smart city surveillance. Identifying miniature components on a factory floor. These are the critical challenges facing modern computer vision—where small object detection (SOD) isn’t just a technical hurdle, but a make-or-break factor for safety, automation, and intelligence.
Despite decades of progress, most deep learning models still fail when objects are small, distant, or densely packed. Standard detectors like YOLO, Faster R-CNN, and RetinaNet often miss them entirely due to low resolution, occlusion, and scale variation. The result? Missed detections, false alarms, and unreliable AI systems.
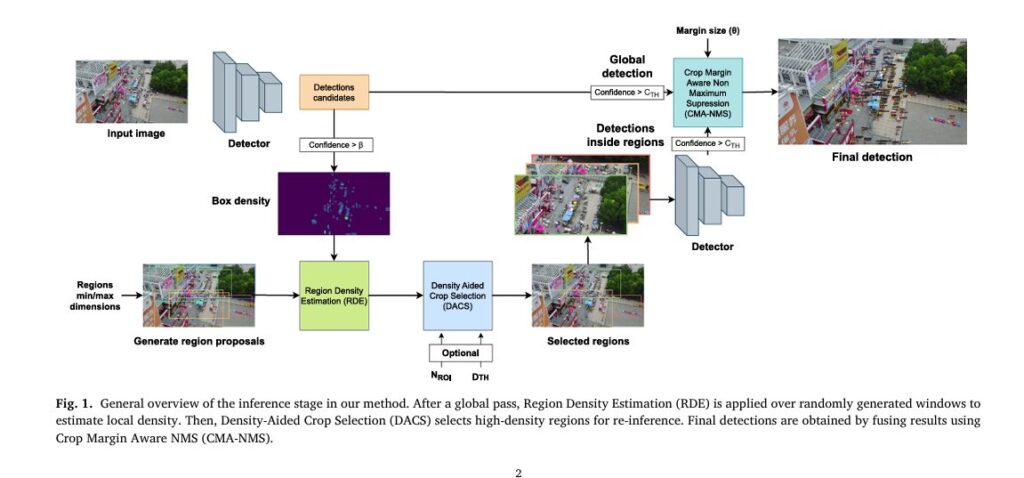
But a new, lightweight, and revolutionary solution is changing the game: DAHI (Density-Aided Hyper Inference)—a fast, efficient, and plug-and-play framework that dramatically improves detection accuracy without retraining or adding complex auxiliary networks.
In this article, we’ll dive deep into 7 powerful breakthroughs that make DAHI a paradigm shift in large-scene object detection—and why it outperforms even the most advanced multi-inference strategies.
1. The Hidden Problem: Why Small Objects Are So Hard to Detect
Small objects—often less than 32×32 pixels—pose unique challenges:
- Low resolution: Few pixels mean weak feature representation.
- Feature suppression: Downsampling in CNNs erases critical details.
- Occlusion and crowding: Objects cluster together, leading to missed detections.
- Scale variation: A single image may contain objects from 10px to 1000px.
Traditional approaches like sliding windows or uniform tiling increase computational cost without guaranteeing better results. Many rely on expensive auxiliary networks or clustering algorithms that add latency and complexity.
But what if you could intelligently focus your inference only on the most promising regions—without any extra training?
That’s exactly what DAHI delivers.
2. DAHI: A Lightweight, Detector-Agnostic Revolution
DAHI stands for Density-Aided Hyper Inference, and it’s not a new detector—it’s a smart inference strategy that works with any existing model:
- ✅ No retraining required
- ✅ Works with YOLO, Faster R-CNN, RetinaNet, DETR, etc.
- ✅ Adds minimal computational overhead
- ✅ Improves accuracy and recall—especially for small objects
The framework consists of three core components:
- Region Density Estimation (RDE)
- Density-Aided Crop Selection (DACS)
- Crop Margin Aware NMS (CMA-NMS)
Together, they form a fast, efficient, and highly effective pipeline that boosts performance where it matters most.
3. Breakthrough #1: Spatial Clustering Reveals the Key
Before designing DAHI, the researchers analyzed four major aerial datasets: VisDrone, UAVDT, SODA-D, and AI-TOD.
They computed the Chebyshev distance between ground truth bounding boxes and found a striking pattern:
85% to 92% of objects lie within a normalized distance of 0.5 from their nearest neighbors.
This means: small objects are not randomly scattered—they cluster.
🔍 Insight: If one small object is detected, there’s a high chance others are nearby and being missed.
This clustering bias is the foundation of DAHI’s intelligence. Instead of blindly slicing the image, DAHI uses density estimation to guide where to look next.
4. Breakthrough #2: Region Density Estimation (RDE)
After an initial global inference pass with a low confidence threshold (β), DAHI generates a set of random candidate regions across the image.
Each region’s density score is calculated based on how many initial detections fall inside it—weighted by overlap.
The density function is defined as:
$$ D(r_i, B) = A(r_i) \cdot \lambda \sum_{b_j \in B} M(r_i, b_j) $$Where:
- ri : candidate region
- B : set of initial detections
- bj : j-th detected bounding box
- A(ri) : area of region ri
- λ : regularization constant (e.g., 0.001)
- M(ri , bj) : overlap measure
The overlap measure is:
\[ M(r_i, b_j) = \begin{cases} \frac{|b_j|}{|r_i \cap b_j|}, & \text{if } \frac{|b_j|}{|r_i \cap b_j|} \geq 0.5 \\ 0, & \text{otherwise} \end{cases} \]This ensures only significant overlaps contribute, reducing noise.
Regions with high density scores are prioritized for re-inference—meaning the system focuses on areas most likely to contain overlooked small objects.
5. Breakthrough #3: Density-Aided Crop Selection (DACS)
Once regions are scored, DACS selects the most informative ones while minimizing overlap.
Inspired by Non-Maximum Suppression (NMS), DACS uses Intersection over Smaller (IoS) to filter overlapping regions.
DACS Algorithm (Simplified)
Sort regions by density score (descending)
ROI = empty list
While regions remain and ROI < NROI and coverage < CDTH:
Select highest-scoring region r1
Add r1 to ROI
Remove all regions with IoS(r1, ri) > τKey parameters:
- NROI: Max number of crops (e.g., 2–3)
- CDTH: Coverage threshold (e.g., 80% of initial detections covered)
- τ: Overlap threshold (e.g., 0.25)
This ensures:
- ✅ High information gain
- ✅ Low redundancy
- ✅ Controlled computational cost
Unlike fixed-grid tiling (e.g., 2×2 or 3×3), DAHI adapts to scene content—focusing only where needed.
6. Breakthrough #4: Crop Margin Aware NMS (CMA-NMS)
One major flaw in multi-inference systems: duplicate detections at crop boundaries.
When a small object is split across two crops, it may be partially detected in one and fully in another. Standard NMS fails to merge them because the IoU is low.
DAHI solves this with CMA-NMS, a smarter suppression rule.
How CMA-NMS Works
For each detection:
- If it’s near the edge of its crop (within θ pixels), it’s flagged.
- When comparing detections from different crops:
- If one is flagged, use IoS with a higher threshold (e.g., 0.9)
- Otherwise, use standard IoU (e.g., 0.65)
The margin test MT(bi, Ci) checks if any box coordinate is within θ pixels of the crop border:
\[ MT(b_i, C_i) = \begin{cases} \text{True}, & \text{if } b_{x1} \in [C_{x1}+\theta,\, C_{x2}-\theta] \\ \text{True}, & \text{if } b_{y1} \in [C_{y1}+\theta,\, C_{y2}-\theta] \\ \text{True}, & \text{if } b_{x2} \in [C_{x1}+\theta,\, C_{x2}-\theta] \\ \text{True}, & \text{if } b_{y2} \in [C_{y1}+\theta,\, C_{y2}-\theta] \\ \text{False}, & \text{otherwise} \end{cases} \]This eliminates false positives caused by boundary artifacts—a common failure in other methods.
7. Breakthrough #5: Superior Performance with Minimal Overhead
DAHI was tested on VisDrone, UAVDT, and SODA-D using five different detectors:
- Faster R-CNN
- FSAF
- RetinaNet
- GFL
- YOLOv10
Key Results:
| DATASET | METHOD | AP50 | #INFERENCE | TOTAL TIME (MS) |
|---|---|---|---|---|
| VisDrone | Baseline | 42.44 | 1.0 | 42.74 |
| SAHI (5.19 inferences) | 49.43 | 5.19 | 174.69 | |
| DAHI | 50.10 | 2.73 | 98.75 |
DAHI achieves higher accuracy with fewer inferences than uniform cropping or SAHI.
On SODA-D, DAHI with YOLOv10 achieved 65.5 AP50—outperforming Uniform Cropping (63.7) while being 72.3% faster.
Breakthrough #6: Ablation Study Proves Each Component Matters
An ablation study confirmed the value of each DAHI module:
| METHOD | AP50 (VISDRONE) | #INFERENCE |
|---|---|---|
| Random Patches | 46.77 | 2.91 |
| Meanshift Clustering | 44.65 | 2.04 |
| DAHI (Full) | 50.10 | 2.73 |
| DAHI (w/ Standard NMS) | 49.53 | 2.73 |
- RDE + DACS improves recall by focusing on high-density regions.
- CMA-NMS adds +0.6 AP50 by eliminating boundary duplicates.
Even when combined with Uniform Cropping, CMA-NMS boosts AP50 by 0.7–1.7 points, proving its broad applicability.
Breakthrough #7: The Perfect Accuracy-Speed Trade-Off
Most multi-inference methods sacrifice speed for accuracy. DAHI breaks this trade-off.
| METHOD | SPEED | ACCURACY | FLEXIBILITY |
|---|---|---|---|
| DAHI | ✅ Fast (2–3 inferences) | ✅ High (Top-tier AP50) | ✅ Plug-and-play |
| Uniform Cropping | ❌ Slow (5+ inferences) | ✅ High | ❌ Fixed grid |
| CRENet/GLSAN | ❌ Very slow (30–100ms extra) | ✅ Medium | ❌ Needs training |
| Baseline | ✅ Fast | ❌ Low (misses small objects) | ✅ Simple |
DAHI’s optional parameters (NROI, CDTH) allow tuning for real-time applications:
- Set NROI=1 for ultra-fast inference
- Use CDTH for early termination when coverage is sufficient
Real-World Applications: Where DAHI Shines
🛰️ Aerial Surveillance (Drones & UAVs)
- Detect vehicles, people, and assets in high-altitude imagery.
- DAHI excels on VisDrone and UAVDT, where objects are tiny and clustered.
🚗 Autonomous Driving
- Spot small obstacles, traffic signs, and pedestrians in wide-angle dashcam footage.
- SODA-D results prove DAHI’s effectiveness in driving scenarios.
🏭 Industrial Inspection
- Identify tiny defects or components on production lines.
- Works with high-resolution images without slowing down.
🌆 Smart City Monitoring
- Scale to large scenes with hundreds of small objects.
- Low-latency inference enables real-time analytics.
Why Other Methods Fail (And DAHI Succeeds)
| METHOD | WHY IT FAILS | HOW DAHI FIXES IT |
|---|---|---|
| Uniform Cropping | Wastes compute on empty regions | Usesdensity to guide crops |
| CRENet/GLSAN | Needs extra networks & training | No retraining; uses detector output |
| Random Crops | Low recall; inefficient | RDE prioritizes informative regions |
| Standard NMS | Fails at crop boundaries | CMA-NMS handles edge duplicates |
DAHI is not just another slicing method—it’s a smarter, data-driven approach that learns from the detector’s own output.
How to Use DAHI in Your Project
DAHI is easy to integrate:
- Train your detector with slicing finetuning (as in SAHI).
- At inference:
- Run global detection with low threshold (β).
- Generate random crops; score with RDE.
- Select top regions with DACS.
- Re-infer and merge with CMA-NMS.
No new training. No extra networks. Just plug-and-play enhancement.
Final Verdict: DAHI Is the Future of Efficient Small Object Detection
While most methods fail due to complexity, latency, or poor recall, DAHI succeeds by being:
- Revolutionary in its use of density estimation
- Fast with only 2–3 inferences
- Accurate across multiple benchmarks
- Flexible and detector-agnostic
It’s not an overstatement to say that DAHI redefines what’s possible in large-scene object detection—especially for edge devices, drones, and real-time systems.
If you’re Interested in defence models against Federated Learning Attacks , you may also find this article helpful: 7 Shocking Federated Learning Attacks That Could Destroy Your Network
Call to Action: Upgrade Your Detection Pipeline Today
Are you tired of missed detections and slow inference in your computer vision projects?
Stop wasting compute on blind tiling. Start using intelligence.
👉 Download the DAHI implementation and integrate it with your YOLO, Faster R-CNN, or RetinaNet model today.
Boost your AP50 by up to 8 points—without retraining.
Cut inference time by 50% compared to uniform slicing.
Deploy smarter, faster, and more reliable AI.
Join the revolution in small object detection—before your competitors do.
Below is the complete, end-to-end Python code for the Density Aided Hyper Inference (DAHI) model, as described in the research paper.
import numpy as np
import cv2
# --- Pretrained Model Integration ---
# To use the real YOLOv10 model, you need to install the ultralytics library:
# pip install ultralytics
try:
from ultralytics import YOLO
ULTRALYTICS_AVAILABLE = True
except ImportError:
ULTRALYTICS_AVAILABLE = False
class YoloV10Detector:
"""
A wrapper for the YOLOv10 model from the 'ultralytics' library to be
used as a detector within the DAHI framework.
"""
def __init__(self, model_path='yolov10n.pt'):
"""
Initializes the YOLOv10 detector.
Args:
model_path (str): The path to the pretrained YOLO model file.
It will be downloaded automatically if not present.
"""
if not ULTRALYTICS_AVAILABLE:
raise ImportError("The 'ultralytics' library is required to use the YOLOv10 detector. Please install it with 'pip install ultralytics'")
self.model = YOLO(model_path)
print(f"YOLOv10 model ('{model_path}') loaded successfully.")
def detect(self, image, confidence_threshold):
"""
Performs object detection on an image.
Args:
image (np.ndarray): The input image.
confidence_threshold (float): The confidence threshold for filtering detections.
Returns:
np.ndarray: An array of detections in [x1, y1, x2, y2, confidence] format.
"""
# Perform inference
results = self.model(image, conf=confidence_threshold, verbose=False)
# Extract bounding boxes and confidences
boxes = results[0].boxes.xyxyn.cpu().numpy() # Normalized [x1, y1, x2, y2]
confidences = results[0].boxes.conf.cpu().numpy()
# Denormalize coordinates
h, w, _ = image.shape
boxes[:, 0] *= w
boxes[:, 1] *= h
boxes[:, 2] *= w
boxes[:, 3] *= h
# Combine into the required format [x1, y1, x2, y2, confidence]
if len(boxes) > 0:
detections = np.hstack((boxes, confidences[:, np.newaxis]))
return detections
else:
return np.array([])
class DummyDetector:
"""A dummy detector that returns random bounding boxes for demonstration."""
def detect(self, image, confidence_threshold):
height, width, _ = image.shape
num_detections = np.random.randint(5, 20)
detections = []
for _ in range(num_detections):
x1 = np.random.randint(0, width - 50)
y1 = np.random.randint(0, height - 50)
x2 = x1 + np.random.randint(20, 100)
y2 = y1 + np.random.randint(20, 100)
confidence = np.random.rand()
if confidence >= confidence_threshold:
detections.append([x1, y1, x2, y2, confidence])
return np.array(detections)
class DAHI:
"""
Implementation of the Density Aided Hyper Inference (DAHI) model.
"""
def __init__(self, model_name='yolov10', confidence_threshold=0.5, rde_confidence_threshold=0.1,
max_regions=10, coverage_threshold=0.9, cma_nms_threshold=0.5,
margin_test_threshold=0.9, margin_size=10):
"""
Initializes the DAHI model with a base detector and configuration parameters.
"""
self.detector = self._load_detector(model_name)
self.confidence_threshold = confidence_threshold
self.rde_confidence_threshold = rde_confidence_threshold
self.max_regions = max_regions
self.coverage_threshold = coverage_threshold
self.cma_nms_threshold = cma_nms_threshold
self.margin_test_threshold = margin_test_threshold
self.margin_size = margin_size
def _load_detector(self, model_name):
"""Loads a pretrained detector model based on the model name."""
if model_name == 'yolov10':
try:
return YoloV10Detector()
except ImportError as e:
print(f"WARNING: {e}. Falling back to DummyDetector.")
return DummyDetector()
elif model_name == 'dummy':
print("Loading DummyDetector for demonstration.")
return DummyDetector()
else:
raise ValueError(f"Model '{model_name}' is not supported. Choose from 'yolov10' or 'dummy'.")
def infer(self, image):
"""
Performs inference on an image using the DAHI pipeline.
"""
# 1. Global Detection
initial_detections = self.detector.detect(image, self.rde_confidence_threshold)
# 2. Candidate Region Sampling
candidate_regions = self._generate_candidate_regions(image)
# 3. Region Density Estimation (RDE)
region_densities = self._calculate_region_densities(candidate_regions, initial_detections)
# 4. Density-Aided Crop Selection (DACS)
selected_regions = self._select_regions_dacs(candidate_regions, region_densities)
# 5. Regional Inference
all_detections = list(initial_detections) if len(initial_detections) > 0 else []
for region in selected_regions:
x1, y1, x2, y2 = [int(c) for c in region]
cropped_image = image[y1:y2, x1:x2]
if cropped_image.size == 0: continue
regional_detections = self.detector.detect(cropped_image, self.confidence_threshold)
if len(regional_detections) > 0:
for det in regional_detections:
det[0] += x1
det[1] += y1
det[2] += x1
det[3] += y1
all_detections.extend(regional_detections)
# 6. Crop Margin Aware NMS (CMA-NMS)
if not all_detections:
return np.array([])
final_detections = self._cma_nms(all_detections, selected_regions)
return np.array(final_detections)
def _generate_candidate_regions(self, image, num_regions=100, min_size=300, max_size=600):
"""
Generates random candidate regions for RDE.
"""
height, width, _ = image.shape
regions = []
for _ in range(num_regions):
actual_max_w = min(max_size, width)
actual_max_h = min(max_size, height)
if min_size >= actual_max_w or min_size >= actual_max_h:
continue
w = np.random.randint(min_size, actual_max_w)
h = np.random.randint(min_size, actual_max_h)
x1 = np.random.randint(0, width - w)
y1 = np.random.randint(0, height - h)
regions.append([x1, y1, x1 + w, y1 + h])
return np.array(regions)
def _calculate_region_densities(self, regions, detections, lambda_reg=0.001):
"""
Calculates the density of detections for each candidate region.
"""
densities = []
if len(detections) == 0:
return np.zeros(len(regions))
for region in regions:
rx1, ry1, rx2, ry2 = region
region_area = (rx2 - rx1) * (ry2 - ry1)
density = 0
for det in detections:
dx1, dy1, dx2, dy2, _ = det
inter_x1 = max(rx1, dx1)
inter_y1 = max(ry1, dy1)
inter_x2 = min(rx2, dx2)
inter_y2 = min(ry2, dy2)
inter_area = max(0, inter_x2 - inter_x1) * max(0, inter_y2 - inter_y1)
det_area = (dx2 - dx1) * (dy2 - dy1)
if det_area > 0 and inter_area / det_area >= 0.5:
density += inter_area / det_area
densities.append(density / (region_area * lambda_reg) if region_area > 0 else 0)
return np.array(densities)
def _select_regions_dacs(self, regions, densities, ios_threshold=0.25):
"""
Selects regions using the DACS algorithm.
"""
if len(regions) == 0:
return []
non_zero_indices = np.where(densities > 0)[0]
if len(non_zero_indices) == 0:
return []
regions = regions[non_zero_indices]
densities = densities[non_zero_indices]
sorted_indices = np.argsort(densities)[::-1]
selected_regions = []
indices_to_process = list(range(len(sorted_indices)))
while len(indices_to_process) > 0 and len(selected_regions) < self.max_regions:
best_local_idx = indices_to_process.pop(0)
best_original_idx = sorted_indices[best_local_idx]
best_region = regions[best_original_idx]
selected_regions.append(best_region)
remaining_indices = []
for local_idx in indices_to_process:
original_idx = sorted_indices[local_idx]
region_to_check = regions[original_idx]
ios = self._calculate_ios(best_region, region_to_check)
if ios <= ios_threshold:
remaining_indices.append(local_idx)
indices_to_process = remaining_indices
return selected_regions
def _cma_nms(self, detections, regions):
"""
Performs Crop Margin Aware NMS on a set of detections.
"""
detection_origins = self._assign_detection_origins(detections, regions)
sorted_indices = sorted(range(len(detections)), key=lambda k: detections[k][4], reverse=True)
final_detections = []
while len(sorted_indices) > 0:
best_idx = sorted_indices.pop(0)
best_det = detections[best_idx]
best_origin = detection_origins[best_idx]
final_detections.append(best_det)
remaining_indices = []
for idx in sorted_indices:
det = detections[idx]
origin = detection_origins[idx]
is_margin_test = False
if best_origin != origin and origin != -1:
crop = regions[origin]
if self._margin_test(det, crop):
is_margin_test = True
if is_margin_test:
ios = self._calculate_ios(best_det, det)
if ios <= self.margin_test_threshold:
remaining_indices.append(idx)
else:
iou = self._calculate_iou(best_det, det)
if iou <= self.cma_nms_threshold:
remaining_indices.append(idx)
sorted_indices = remaining_indices
return final_detections
def _assign_detection_origins(self, detections, regions):
"""Assigns an origin index to each detection."""
origins = []
for det in detections:
cx = (det[0] + det[2]) / 2
cy = (det[1] + det[3]) / 2
assigned_origin = -1
for i, region in enumerate(regions):
if region[0] <= cx < region[2] and region[1] <= cy < region[3]:
assigned_origin = i
break
origins.append(assigned_origin)
return origins
def _margin_test(self, detection, crop):
"""Checks if a detection is within the margin of a crop."""
x1, y1, x2, y2, _ = detection
cx1, cy1, cx2, cy2 = crop
return not (cx1 + self.margin_size <= x1 and
cy1 + self.margin_size <= y1 and
x2 <= cx2 - self.margin_size and
y2 <= cy2 - self.margin_size)
def _calculate_iou(self, box1, box2):
"""Calculates Intersection over Union."""
x1 = max(box1[0], box2[0])
y1 = max(box1[1], box2[1])
x2 = min(box1[2], box2[2])
y2 = min(box1[3], box2[3])
inter_area = max(0, x2 - x1) * max(0, y2 - y1)
area1 = (box1[2] - box1[0]) * (box1[3] - box1[1])
area2 = (box2[2] - box2[0]) * (box2[3] - box2[1])
union_area = area1 + area2 - inter_area
return inter_area / union_area if union_area > 0 else 0
def _calculate_ios(self, box1, box2):
"""Calculates Intersection over Smaller."""
x1 = max(box1[0], box2[0])
y1 = max(box1[1], box2[1])
x2 = min(box1[2], box2[2])
y2 = min(box1[3], box2[3])
inter_area = max(0, x2 - x1) * max(0, y2 - y1)
area1 = (box1[2] - box1[0]) * (box1[3] - box1[1])
area2 = (box2[2] - box2[0]) * (box2[3] - box2[1])
smaller_area = min(area1, area2)
return inter_area / smaller_area if smaller_area > 0 else 0
# Example Usage
if __name__ == '__main__':
# --- Select the model to use ---
# Change to 'dummy' if you don't have 'ultralytics' installed.
MODEL_TO_USE = 'yolov10'
print(f"--- Initializing DAHI with '{MODEL_TO_USE}' model ---")
dahi_model = DAHI(model_name=MODEL_TO_USE)
# Create a dummy image for testing
dummy_image = np.zeros((1080, 1920, 3), dtype=np.uint8)
# Add some "objects" to the dummy image for the detector to find
cv2.rectangle(dummy_image, (100, 100), (250, 250), (255, 0, 0), -1) # A person
cv2.rectangle(dummy_image, (120, 120), (280, 280), (255, 0, 0), -1) # Another person
cv2.rectangle(dummy_image, (800, 400), (1100, 600), (0, 0, 255), -1) # A car
# Perform inference
print("\n--- Starting DAHI inference ---")
final_detections = dahi_model.infer(dummy_image)
print(f"--- DAHI inference complete ---")
print(f"\nDAHI found {len(final_detections)} objects.")
# Visualize the detections
for x1, y1, x2, y2, conf in final_detections:
cv2.rectangle(dummy_image, (int(x1), int(y1)), (int(x2), int(y2)), (0, 255, 0), 2)
cv2.putText(dummy_image, f"{conf:.2f}", (int(x1), int(y1) - 10),
cv2.FONT_HERSHEY_SIMPLEX, 0.5, (0, 255, 0), 2)
# Save the image with detections to a file
output_filename = "dahi_yolo_detections.jpg"
cv2.imwrite(output_filename, dummy_image)
print(f"Saved detection results to {output_filename}")

Pingback: 7 Shocking Vulnerabilities in AI Watermarking: The Hidden Threat of Unified Spoofing & Scrubbing Attacks (And How to Fix It) - aitrendblend.com
Pingback: 5 Shocking Mistakes in Knowledge Distillation (And the Brilliant Framework KD2M That Fixes Them) - aitrendblend.com
Pingback: 7 Shocking Ways Integrated Gradients BOOST Knowledge Distillation - aitrendblend.com