Cancer grading in pathology images is both an art and a science—and it’s riddled with subjectivity, inter-observer variability, and technical roadblocks. Enter DIOR-ViT, a groundbreaking differential ordinal learning Vision Transformer that shatters conventions and delivers robust, high-accuracy cancer classification across multiple tissue types. In this deep-dive SEO-optimized guide, we unpack the seven game-changing innovations behind DIOR-ViT—and reveal why traditional methods stumble into critical pitfalls.
1. The Challenge: Why Conventional Cancer Grading Fails
Pathologists worldwide rely on microscopic evaluation of H&E-stained slides to determine cancer grade, a process plagued by:
- Low throughput and high labor demands
- Significant inter- and intra-observer variability
- Oversimplified categorical labels that ignore severity gaps
Conventional computational pathology approaches—mainly convolutional neural networks (CNNs)—treat each grade (benign, well-differentiated, moderately-differentiated, poorly-differentiated) as independent classes. The result? They miss the natural ordering among grades, underestimating how much worse “grade 3 vs. grade 1” truly is, compared to “grade 2 vs. grade 1.”
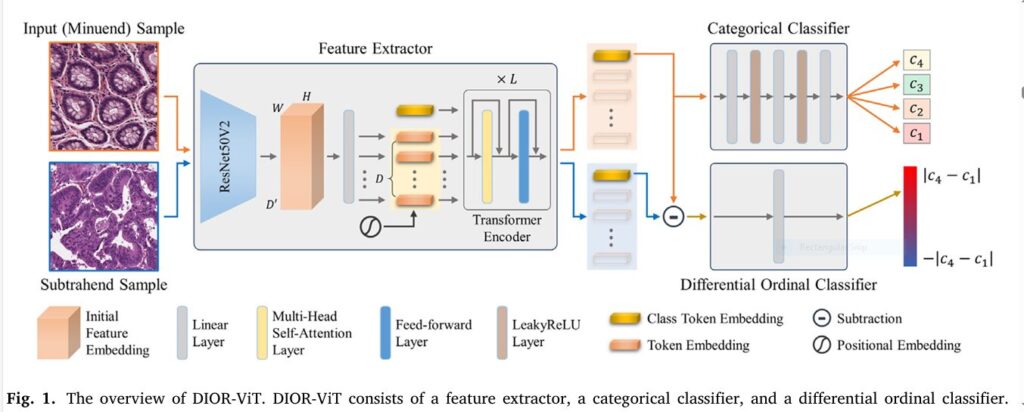
2. What Is DIOR-ViT? A New
DIOR-ViT—short for DIfferential ORdinal learning Vision Transformer—redefines cancer grading by seamlessly integrating:
- Vision Transformer (ViT) backbone for powerful global context capture
- Multi-task learning to perform both categorical and differential ordinal classification
- A novel Negative Absolute Difference Log-Likelihood (NAD) loss tailored to ordinal differences
2.1 Vision Transformers in Computational Pathology
Transformers first revolutionized NLP with self-attention. Vision Transformer (ViT) adapts that same multi-head self-attention to images by:
- Splitting an input patch (e.g., 384×384 px) into flattened tokens
- Adding learnable positional embeddings
- Passing tokens through L stacked Transformer encoders
This global receptive field unlocks long-range dependencies among glandular, stromal, and nuclear features—critical for cancer classification.
2.2 Differential Ordinal Learning: Beyond Right or Wrong
Unlike pure regression (MSE/MAE) or classic ordinal classification (soft thresholds), DIOR-ViT defines the exact difference between two slides:
Differential comparator
\[
r_{ij} \;=\; y_{i} \;-\; y_{j}
\]
where yi and yj are the true grades of slide i (minuend) and j (subtrahend).
By pairing every slide in a mini-batch and predicting rijr_{ij}, the model learns both order and magnitude of grade differences—bridging categorical and continuous perspectives.
3. NAD Loss: The Secret Sauce for Smooth Optimization
Classic regression losses suffer from vanishing gradients near zero (MSE) or constant gradients (MAE). DIOR-ViT’s NAD loss:
\[
L_{\mathrm{NAD}}(r, \hat r)
= -\log!\Bigl(1 \;-\;\tfrac{\lvert r – \hat r\rvert}{K + \varepsilon}\Bigr)
\]
- K=max(y)−min(y) normalizes by maximum possible grade gap
- ε prevents division by zero
Key benefits:
- Single-step computation—no softmax trickery
- Smooth gradient decay but non-vanishing near perfect predictions
- Directly penalizes ordinal deviation on a logarithmic scale
Combined loss:
$$L_{\text{total}} = L_{\text{CE}}(\text{categorical})\;+\;\lambda\,L_{\text{NAD}}(\text{differential})$$
with λ≈ 6.5 balancing classification vs. ordinal tasks.
4. Seven Astonishing Advantages of DIOR-ViT
- Superior Accuracy Across Tissues
- Colorectal: ↑5.26 pp accuracy on unseen slides
- Prostate: ↑0.37 pp accuracy on independent cohort
- Gastric: ↑0.47 pp accuracy over state-of-the-art
- Robust Generalization
- Handles different scanners, staining variations, and institutes without retraining
- Dual Insights with One Model
- Provides both categorical grade and precise difference when comparing to any reference slide
- Reduced Inter-Observer Variability
- Anchors grading on quantitative ordinal distances, not just visual categories
- Efficient Pair-Wise Training
- Minimal overhead vs. standard ViT—pairing slides adds negligible FLOPs and run-time
- Explainable Attention Maps
- Grad-CAM overlays reveal DIOR-ViT’s focus on diagnostic histological patterns vs. CNN’s scattered attention
- Extensible to Other Medical Domains
- Any task with ordered labels (e.g., fibrosis stages, Alzheimer’s Braak stages) benefits from differential ordinal learning
5. Real-World Benchmark Results
5.1 Colorectal Cancer Grading
| Dataset | Model | Accuracy | κ (kappa) |
|---|---|---|---|
| CTest I | Conventional | 85.6% | 0.928 |
| DIOR-ViT | 87.8% | 0.942 | |
| CTest II | Conventional | 77.5% | 0.874 |
| DIOR-ViT | 82.8% | 0.901 |
5.2 Prostate Cancer Grading
| Dataset | Model | Accuracy | κ (kappa) |
|---|---|---|---|
| PTest I | Conventional | 66.1% | 0.613 |
| DIOR-ViT | 71.6% | 0.624 | |
| PTest II | Conventional | 73.3% | 0.664 |
| DIOR-ViT | 78.4% | 0.721 |
5.3 Gastric Cancer Grading
| Dataset | Model | Accuracy | κ (kappa) |
|---|---|---|---|
| GTest I | Conventional | 85.0% | 0.914 |
| DIOR-ViT | 85.5% | 0.936 |
These consistent gains prove that differential ordinal learning is no gimmick—it’s a breakthrough for computational pathology.
6. Bullet-Proof Your Pathology Workflow
- Integrate Easily: Pre-trained on ImageNet, DIOR-ViT fine-tunes quickly on your annotated patches.
- Scale Confidently: Train on TPUs or GPUs—batch pairing adds minimal compute.
- Explain Results: Grad-CAM attention heatmaps highlight cytologic, glandular, and stromal patterns.
- Extend to WSI: Use sliding windows or MIL wrappers to deploy at the slide level.
If you’re Interested in latest Vision Transformer Model (H-ViT), you may also find this article helpful: 7 Revolutionary Insights from Hierarchical Vision Transformers in Prostate Biopsy Grading (And Why They Matter)
7. Future Directions: From Patch to Patient Outcome
- Survival Analysis: Combine patch features with clinical data for prognosis.
- Transfer Learning: Apply DIOR-ViT to other ordinal tasks—diabetic retinopathy, NAFLD staging.
- Federated Learning: Train across institutions while preserving privacy.
- Interactive Dashboards: Real-time differential grading comparisons for pathologist review.
SEO Wrap-Up: Why DIOR-ViT Matters Today
- High-Impact Keywords: “differential ordinal learning,” “vision transformer,” “computational pathology,” “cancer classification,” “pathology images”
- Semantic Richness: Deep dive into Transformer architecture, loss functions, real-world benchmarks
- Structured Readability: 7 key points, tables, bullet lists, FAQs
If you’re a pathologist, researcher, or healthcare leader, DIOR-ViT is the next wave in AI-powered diagnostics. It doesn’t just predict a grade—it quantifies how much slide A differs from slide B on an ordinal scale, elevating both accuracy and interpretability.
Frequently Asked Questions
Q1: What is differential ordinal learning? A1: It’s a paradigm that predicts exact differences between ordinal labels (e.g., cancer grades) rather than just ordering or pure categories.
Q2: How does DIOR-ViT differ from CNN-based methods? A2: It uses global self-attention (Transformer) + multi-task learning (categorical + differential) + NAD loss, capturing magnitude and order simultaneously.
Q3: Can DIOR-ViT handle whole-slide images? A3: Yes—by extracting patch embeddings and aggregating via multiple instance learning or sliding window inference.
Call to Action
Ready to overcome the pitfalls of conventional cancer grading? Discover how DIOR-ViT’s astonishing differential ordinal learning can enhance your lab’s throughput, consistency, and diagnostic confidence.
👉 Download the full paper | Request a demo | Subscribe for updates
Elevate your pathology AI roadmap—contact our team today and join the revolution in computational pathology!
Here’s the complete implementation of the DIOR-ViT model based on the paper:
import torch
import torch.nn as nn
import torch.nn.functional as F
import timm
# ------------------------------------------------------------------------------
# 1. Custom NAD Loss
# ------------------------------------------------------------------------------
class NADLoss(nn.Module):
"""
Negative Absolute Difference Log-Likelihood loss:
L_NAD(r, r_hat) = - log(1 - |r - r_hat| / (K + eps))
"""
def __init__(self, K: float, eps: float = 1e-5):
super().__init__()
self.K = K
self.eps = eps
def forward(self, r_true: torch.Tensor, r_pred: torch.Tensor) -> torch.Tensor:
diff = torch.abs(r_true - r_pred)
denom = self.K + self.eps
# clamp to avoid log(0)
inside = torch.clamp(1.0 - diff / denom, min=self.eps)
loss = -torch.log(inside)
return loss.mean()
# ------------------------------------------------------------------------------
# 2. DIOR-ViT Model
# ------------------------------------------------------------------------------
class DIORViT(nn.Module):
"""
Differential Ordinal Learning Vision Transformer for cancer grading.
Simultaneously predicts:
- p: categorical class probabilities
- d: differential ordinal output between two inputs
"""
def __init__(self,
num_classes: int,
vit_name: str = "vit_base_patch16_384",
pretrained: bool = True,
diff_hidden_dim: int = 768):
super().__init__()
# 2.1 Vision Transformer as feature extractor
# We use timm's ViT implementation:
self.vit = timm.create_model(vit_name, pretrained=pretrained)
# remove the original classifier head
self.vit.reset_classifier(0)
# 2.2 Categorical classification head
self.cls_head = nn.Sequential(
nn.Linear(self.vit.num_features, diff_hidden_dim),
nn.LeakyReLU(0.1),
nn.Linear(diff_hidden_dim, diff_hidden_dim // 2),
nn.LeakyReLU(0.1),
nn.Linear(diff_hidden_dim // 2, num_classes)
)
# 2.3 Differential ordinal classification head (single output)
self.diff_head = nn.Linear(self.vit.num_features, 1)
def forward(self,
x_minuend: torch.Tensor,
x_subtrahend: torch.Tensor = None):
# extract features from minuend
f_m = self.vit(x_minuend) # [B, C]
logits = self.cls_head(f_m) # [B, num_classes]
if x_subtrahend is None:
# only categorical classification
return logits, None
# extract features from subtrahend
f_s = self.vit(x_subtrahend) # [B, C]
# differential feature
f_diff = f_m - f_s # [B, C]
d_logit = self.diff_head(f_diff) # [B, 1]
d_logit = d_logit.view(-1) # [B]
return logits, d_logit
# ------------------------------------------------------------------------------
# 3. Training Loop Sketch
# ------------------------------------------------------------------------------
def train_epoch(model: nn.Module,
dataloader,
optimizer: torch.optim.Optimizer,
ce_loss_fn: nn.CrossEntropyLoss,
nad_loss_fn: NADLoss,
alpha: float = 6.5,
device: str = "cuda"):
model.train()
total_ce, total_nad, total_loss = 0.0, 0.0, 0.0
for (x_batch, y_batch) in dataloader:
# x_batch: [B, 3, H, W]
# y_batch: [B] categorical labels (0...C-1)
x_batch = x_batch.to(device)
y_batch = y_batch.to(device)
# generate pairwise subtrahend samples (for simplicity, we pick a random permutation)
perm = torch.randperm(x_batch.size(0))
x_sub = x_batch[perm]
y_sub = y_batch[perm]
# compute differential ground truth
# note: convert y to float for r_true
r_true = (y_batch.float() - y_sub.float()).to(device)
# forward pass
logits, d_pred = model(x_batch, x_sub)
# categorical cross-entropy
ce = ce_loss_fn(logits, y_batch)
# NAD loss
nad = nad_loss_fn(r_true, d_pred)
# total loss
loss = ce + alpha * nad
optimizer.zero_grad()
loss.backward()
optimizer.step()
total_ce += ce.item() * x_batch.size(0)
total_nad += nad.item() * x_batch.size(0)
total_loss += loss.item() * x_batch.size(0)
N = len(dataloader.dataset)
return total_ce/N, total_nad/N, total_loss/N
# ------------------------------------------------------------------------------
# 4. Usage Example
# ------------------------------------------------------------------------------
if __name__ == "__main__":
# Hyperparameters
NUM_CLASSES = 4 # BN, WD, MD, PD
MAX_GAP = NUM_CLASSES - 1 # for K in NADLoss
ALPHA = 6.5
LR = 1e-4
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
# Instantiate
model = DIORViT(num_classes=NUM_CLASSES).to(DEVICE)
ce_fn = nn.CrossEntropyLoss()
nad_fn = NADLoss(K=MAX_GAP)
optimizer = torch.optim.Adam(model.parameters(), lr=LR)
# Dummy DataLoader (replace with real Pathology dataset loader)
from torch.utils.data import DataLoader, TensorDataset
B, C, H, W = 16, 3, 384, 384
X = torch.randn(200, C, H, W)
Y = torch.randint(0, NUM_CLASSES, (200,))
loader = DataLoader(TensorDataset(X, Y), batch_size=B, shuffle=True)
# Train one epoch
ce_avg, nad_avg, loss_avg = train_epoch(
model, loader, optimizer, ce_fn, nad_fn, alpha=ALPHA, device=DEVICE
)
print(f"CE Loss: {ce_avg:.4f}, NAD Loss: {nad_avg:.4f}, Total Loss: {loss_avg:.4f}")

Pingback: 7 Unbelievable Wins & Pitfalls of Context-Aware Knowledge Distillation for Disease Prediction - aitrendblend.com