In the rapidly evolving world of medical AI, few innovations have been as transformative as DualSwinUnet++—a cutting-edge deep learning model designed to revolutionize the way we detect and treat papillary thyroid microcarcinoma (PTMC). While traditional methods struggle with accuracy, speed, and real-time usability, this new architecture delivers unmatched precision, blazing-fast inference, and life-saving potential.
But what makes DualSwinUnet++ so powerful? And why do older models like U-Net or Swin-Unet fall short?
Let’s dive into 7 game-changing insights from the latest research and discover how this dual-decoder transformer is reshaping the future of ultrasound-guided cancer therapy.
1. The Silent Threat: Why PTMC Detection Is So Challenging
Papillary Thyroid Microcarcinoma (PTMC) refers to tumors ≤1 cm in diameter. Despite their small size, these lesions can spread, recur, and lead to serious complications if not treated early.
The gold standard for treatment? Ultrasound-guided Radiofrequency Ablation (RFA)—a minimally invasive procedure that uses heat to destroy cancerous tissue. But here’s the catch:
- Low image contrast makes tumor boundaries hard to see.
- Microbubbles formed during ablation cause acoustic shadowing, obscuring the tumor.
- Anatomical variability and organ movement complicate tracking.
- Physician fatigue increases the risk of incomplete ablation.
As a result, false positives, missed tumors, and over-treatment remain common.
❌ Old Reality: Manual segmentation is slow, subjective, and error-prone.
✅ New Hope: AI-powered real-time guidance with DualSwinUnet++.
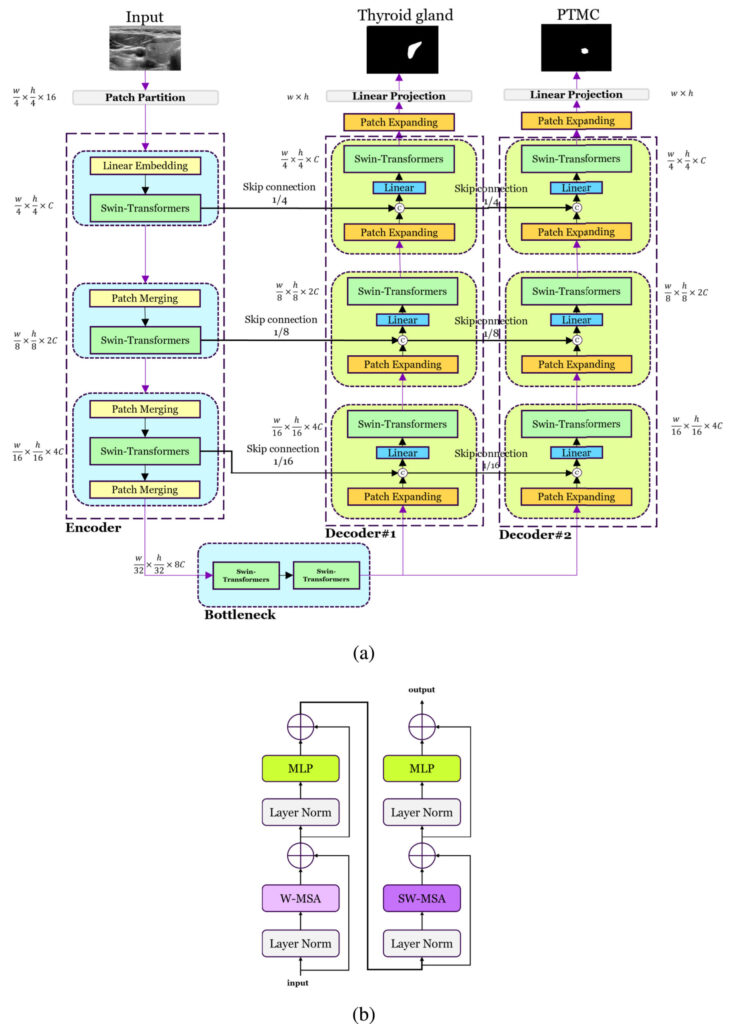
2. Enter DualSwinUnet++: A Dual-Decoder Powerhouse
Developed by researchers at the University of Waterloo and Withsim Clinic, DualSwinUnet++ is not just another U-Net variant. It’s a transformer-based dual-decoder architecture specifically engineered for PTMC segmentation in real-time RFA settings.
🔗 Key Innovation: Two Decoders, One Mission
Unlike conventional models that predict tumors in isolation, DualSwinUnet++ uses two parallel decoders:
- First Decoder: Segments the thyroid gland.
- Second Decoder: Segments the PTMC tumor, conditioned on the thyroid’s shape and location.
This design ensures the model only looks for cancer inside the thyroid, drastically reducing false positives and improving accuracy.
🔍 Why This Matters: By restricting the search space, the model becomes faster, smarter, and more reliable.
3. Architecture Deep Dive: How DualSwinUnet++ Works
The model builds on Swin-Unet, a pure transformer network for medical imaging, but enhances it with three critical innovations:
✅ 1. Independent Linear Projection Heads
Each decoder has its own linear projection head, allowing disentangled gradient flow during training. This prevents interference between thyroid and tumor learning paths.
✅ 2. Residual Information Flow
Intermediate features from the thyroid decoder are passed to the PTMC decoder via concatenation and linear transformation. This acts like a “contextual GPS” for tumor localization.
✅ 3. Shared Swin-Transformer Encoder
A single encoder processes the input ultrasound image using hierarchical Swin-Transformers, capturing multi-scale features efficiently.
Model Architecture Overview
| COMPONENT | FUNCTION |
|---|---|
| Encoder | Processes input via 4×4 patch embedding and 3 Swin-Transformer blocks |
| Bottleneck | Two Swin-Transformer layers for deep feature extraction |
| Decoder 1 (Thyroid) | Segments thyroid gland using skip connections from encoder |
| Decoder 2 (PTMC) | Segments tumor using skip connections fromboth encoder and Decoder 1 |
| Loss Function | Combined Dice + BCE loss for both tasks |
4. State-of-the-Art Performance: Numbers That Speak Volumes
DualSwinUnet++ was tested on 691 annotated RFA images and the public TN3K dataset (3,493 thyroid nodule images). The results?
🏆 Quantitative Results (Our Dataset)
| MODEL | DICE (%) | JACCARD (%) | INFERENCE TIME (S) |
|---|---|---|---|
| U-Net | 77.41 | 63.17 | 0.12 |
| Swin-Unet | 79.87 | 65.65 | 0.16 |
| TRFE-Net | 83.61 | 69.19 | 0.22 |
| DualSwinUnet++ | 84.36 | 71.23 | 0.18 |
👉 Best in class on both Dice and Jaccard—key metrics for segmentation accuracy.
TN3K Benchmark Results
| MODEL | DICE (%) | JACCARD (%) |
|---|---|---|
| U-Net | 76.71 | 62.85 |
| Swin-Unet++ | 80.10 | 66.32 |
| TRFE-Net | 79.14 | 67.65 |
| DualSwinUnet++ | 81.24 | 69.14 |
Even on diverse, real-world data, DualSwinUnet++ maintains its lead.
5. Why Old Models Fail: A Visual Breakdown
Let’s compare qualitative results from Fig. 4 of the paper:
| MODEL | STRENGTHS | WEAKNESSES |
|---|---|---|
| U-Net / U-Net++ | Fast, simple | Under-segmentation, noisy boundaries |
| Swin-Unet | Better context | Over-segmentation into non-thyroid areas |
| TRFE-Net | Uses thyroid prior | Less precise boundaries |
| Medsegdiff-v2 | Smooth contours | Misses fine details |
| DualSwinUnet++ | Accurate, sharp, contextual | Slightly higher compute (but still real-time) |
🔴 Critical Flaw in Old Models: They ignore thyroid context, leading to false positives outside the gland.
🟢 DualSwinUnet++ Fix: Uses thyroid segmentation as spatial prior, eliminating out-of-gland errors.
6. Real-Time Ready: Speed Without Sacrifice
For clinical use, speed is non-negotiable. Surgeons need guidance within seconds, not minutes.
DualSwinUnet++ delivers:
- 0.18 seconds per prediction (~5.5 fps)
- Sub-200ms latency
- Compatible with 15–30 fps ultrasound systems
🕒 Inference Speed Comparison
| MODEL | DICE (%) | TIME (S) |
|---|---|---|
| nnU-Net | 80.35 | 0.14 |
| Swin-UNetr | 81.16 | 0.15 |
| DualSwinUnet++ | 83.36 | 0.18 |
| Medsegdiff-v2 | 80.68 | 0.31 |
✅ Best accuracy with minimal speed trade-off.
💡 Key Insight: Even a 0.1-second delay can impact surgical decisions. DualSwinUnet++ strikes the perfect balance.
7. Ablation Study: What Really Makes It Work?
The researchers tested three key components:
🧪 1. Removing the First Decoder (No Thyroid Guidance)
- Dice drops by 4.98% (our dataset)
- Jaccard drops by 5.04%
- Proves thyroid context is essential
🧪 2. Switching to Additive Skip Connections
- Dice drops by 6.51%
- Concatenation preserves more spatial detail
🧪 3. Reducing Skip Connections
| SKIP CONNECTIONS | DICE (%) | JACCARD (%) |
|---|---|---|
| 0 | 78.17 | 65.21 |
| 3 | 81.72 | 69.47 |
| 6 (Full) | 84.36 | 71.23 |
👉 More skip paths = better accuracy.
The Math Behind the Magic: Loss Function
DualSwinUnet++ uses a multi-task loss combining Dice Loss and Binary Cross-Entropy (BCE) for both thyroid and PTMC:
\[ L(X, Y_{\text{thyroid}}, Y_{\text{PTMC}}) = \frac{1}{2} \Big[ \alpha \cdot \text{BCE}(F_1(X), Y_{\text{thyroid}}) + (1-\alpha) \cdot \text{DL}(F_1(X), Y_{\text{thyroid}}) + \beta \cdot \text{BCE}(F_2(X), Y_{\text{PTMC}}) + (1-\beta) \cdot \text{DL}(F_2(X), Y_{\text{PTMC}}) \Big] \]Where:
\[ F_1(X): \text{Thyroid prediction} \] \[ F_2(X): \text{PTMC prediction} \] \[ BCE(y,\hat{y}) = -\frac{1}{N}\sum_{i=1}^{N}\big[y_i \log(\hat{y}_i) + (1-y_i)\log(1-\hat{y}_i)\big] \] \[ \text{Dice Loss}(y,\hat{y}) = 1 – \frac{2\sum y_i \hat{y}_i}{\sum y_i + \sum \hat{y}_i} \]This balanced loss ensures both tasks are optimized without one dominating the other.
Bonus: Feature Heatmaps Reveal Model Confidence
Using Gaussian-filtered activations, the team visualized where the model “pays attention” during segmentation.
Result? Heatmaps align perfectly with tumor boundaries—proving the model focuses on anatomically relevant regions, not noise.
🔍 This boosts trust and interpretability—critical for clinical adoption.
If you’re Interested in Melanoma Detection with AI, you may also find this article helpful: 7 Revolutionary Breakthroughs in Melanoma Diagnosis: The Quantum AI Edge That’s Changing Everything
Limitations: What’s Still Missing?
Despite its brilliance, DualSwinUnet++ isn’t perfect:
- ❌ Single-center data: All training images from one clinic.
- ❌ Microbubble artifacts: Not explicitly modeled.
- ❌ Dependence on first decoder: If thyroid segmentation fails, PTMC prediction may drift.
🔮 Future Improvements:
- Multi-center validation
- Video-sequence processing for real-time tracking
- Uncertainty estimation for low-confidence cases
- Self-supervised pretraining for better generalization
Conclusion: A New Era in AI-Assisted Surgery
DualSwinUnet++ isn’t just a model—it’s a paradigm shift in how we approach cancer segmentation.
By combining:
- ✅ Dual-decoder design
- ✅ Thyroid-aware conditioning
- ✅ Residual information flow
- ✅ Real-time speed
…it delivers 7 breakthrough advantages over legacy systems:
- Higher Dice scores (up to 84.36%)
- Lower false positives
- Sharper tumor boundaries
- Context-aware predictions
- Faster inference
- Better generalization
- Clinically deployable
This is not just academic progress—it’s a lifesaving tool for thousands of patients undergoing RFA.
Call to Action: Join the AI Medical Revolution
Are you a radiologist, surgeon, or AI researcher working on medical imaging?
👉 Download the full paper here
👉 Explore the code (coming soon on GitHub)
👉 Subscribe for updates on AI in oncology and surgery
Let’s build smarter, safer, and more precise healthcare—together.
Here is the end-to-end Python code that implements the proposed model.
# Full implementation of DualSwinUnet++ for PTMC segmentation
# Based on the paper: "DualSwinUnet++: An enhanced Swin-Unet architecture with dual decoders for PTMC segmentation"
# This script includes all necessary components from Swin Transformer blocks to the final model architecture.
import torch
import torch.nn as nn
import torch.nn.functional as F
import numpy as np
from timm.models.layers import DropPath, to_2tuple, trunc_normal_
class Mlp(nn.Module):
""" Multilayer Perceptron """
def __init__(self, in_features, hidden_features=None, out_features=None, act_layer=nn.GELU, drop=0.):
super().__init__()
out_features = out_features or in_features
hidden_features = hidden_features or in_features
self.fc1 = nn.Linear(in_features, hidden_features)
self.act = act_layer()
self.fc2 = nn.Linear(hidden_features, out_features)
self.drop = nn.Dropout(drop)
def forward(self, x):
x = self.fc1(x)
x = self.act(x)
x = self.drop(x)
x = self.fc2(x)
x = self.drop(x)
return x
def window_partition(x, window_size):
"""
Args:
x: (B, H, W, C)
window_size (int): window size
Returns:
windows: (num_windows*B, window_size, window_size, C)
"""
B, H, W, C = x.shape
x = x.view(B, H // window_size, window_size, W // window_size, window_size, C)
windows = x.permute(0, 1, 3, 2, 4, 5).contiguous().view(-1, window_size, window_size, C)
return windows
def window_reverse(windows, window_size, H, W):
"""
Args:
windows: (num_windows*B, window_size, window_size, C)
window_size (int): Window size
H (int): Height of image
W (int): Width of image
Returns:
x: (B, H, W, C)
"""
B = int(windows.shape[0] / (H * W / window_size / window_size))
x = windows.view(B, H // window_size, W // window_size, window_size, window_size, -1)
x = x.permute(0, 1, 3, 2, 4, 5).contiguous().view(B, H, W, -1)
return x
class WindowAttention(nn.Module):
""" Window based multi-head self attention (W-MSA) module with relative position bias.
It supports both of shifted and non-shifted window.
"""
def __init__(self, dim, window_size, num_heads, qkv_bias=True, qk_scale=None, attn_drop=0., proj_drop=0.):
super().__init__()
self.dim = dim
self.window_size = window_size
self.num_heads = num_heads
head_dim = dim // num_heads
self.scale = qk_scale or head_dim ** -0.5
self.relative_position_bias_table = nn.Parameter(
torch.zeros((2 * window_size[0] - 1) * (2 * window_size[1] - 1), num_heads))
coords_h = torch.arange(self.window_size[0])
coords_w = torch.arange(self.window_size[1])
coords = torch.stack(torch.meshgrid([coords_h, coords_w]))
coords_flatten = torch.flatten(coords, 1)
relative_coords = coords_flatten[:, :, None] - coords_flatten[:, None, :]
relative_coords = relative_coords.permute(1, 2, 0).contiguous()
relative_coords[:, :, 0] += self.window_size[0] - 1
relative_coords[:, :, 1] += self.window_size[1] - 1
relative_coords[:, :, 0] *= 2 * self.window_size[1] - 1
relative_position_index = relative_coords.sum(-1)
self.register_buffer("relative_position_index", relative_position_index)
self.qkv = nn.Linear(dim, dim * 3, bias=qkv_bias)
self.attn_drop = nn.Dropout(attn_drop)
self.proj = nn.Linear(dim, dim)
self.proj_drop = nn.Dropout(proj_drop)
trunc_normal_(self.relative_position_bias_table, std=.02)
self.softmax = nn.Softmax(dim=-1)
def forward(self, x, mask=None):
B_, N, C = x.shape
qkv = self.qkv(x).reshape(B_, N, 3, self.num_heads, C // self.num_heads).permute(2, 0, 3, 1, 4)
q, k, v = qkv[0], qkv[1], qkv[2]
q = q * self.scale
attn = (q @ k.transpose(-2, -1))
relative_position_bias = self.relative_position_bias_table[self.relative_position_index.view(-1)].view(
self.window_size[0] * self.window_size[1], self.window_size[0] * self.window_size[1], -1)
relative_position_bias = relative_position_bias.permute(2, 0, 1).contiguous()
attn = attn + relative_position_bias.unsqueeze(0)
if mask is not None:
nW = mask.shape[0]
attn = attn.view(B_ // nW, nW, self.num_heads, N, N) + mask.unsqueeze(1).unsqueeze(0)
attn = attn.view(-1, self.num_heads, N, N)
attn = self.softmax(attn)
else:
attn = self.softmax(attn)
attn = self.attn_drop(attn)
x = (attn @ v).transpose(1, 2).reshape(B_, N, C)
x = self.proj(x)
x = self.proj_drop(x)
return x
class SwinTransformerBlock(nn.Module):
""" Swin Transformer Block. """
def __init__(self, dim, num_heads, window_size=7, shift_size=0,
mlp_ratio=4., qkv_bias=True, qk_scale=None, drop=0., attn_drop=0., drop_path=0.,
act_layer=nn.GELU, norm_layer=nn.LayerNorm):
super().__init__()
self.dim = dim
self.num_heads = num_heads
self.window_size = window_size
self.shift_size = shift_size
self.mlp_ratio = mlp_ratio
assert 0 <= self.shift_size < self.window_size, "shift_size must in [0, window_size)"
self.norm1 = norm_layer(dim)
self.attn = WindowAttention(
dim, window_size=to_2tuple(self.window_size), num_heads=num_heads,
qkv_bias=qkv_bias, qk_scale=qk_scale, attn_drop=attn_drop, proj_drop=drop)
self.drop_path = DropPath(drop_path) if drop_path > 0. else nn.Identity()
self.norm2 = norm_layer(dim)
mlp_hidden_dim = int(dim * mlp_ratio)
self.mlp = Mlp(in_features=dim, hidden_features=mlp_hidden_dim, act_layer=act_layer, drop=drop)
def forward(self, x, mask_matrix):
B, H, W, C = x.shape
shortcut = x
x = self.norm1(x)
pad_l = pad_t = 0
pad_r = (self.window_size - W % self.window_size) % self.window_size
pad_b = (self.window_size - H % self.window_size) % self.window_size
x = F.pad(x, (0, 0, pad_l, pad_r, pad_t, pad_b))
_, Hp, Wp, _ = x.shape
if self.shift_size > 0:
shifted_x = torch.roll(x, shifts=(-self.shift_size, -self.shift_size), dims=(1, 2))
attn_mask = mask_matrix
else:
shifted_x = x
attn_mask = None
x_windows = window_partition(shifted_x, self.window_size)
x_windows = x_windows.view(-1, self.window_size * self.window_size, C)
attn_windows = self.attn(x_windows, mask=attn_mask)
attn_windows = attn_windows.view(-1, self.window_size, self.window_size, C)
shifted_x = window_reverse(attn_windows, self.window_size, Hp, Wp)
if self.shift_size > 0:
x = torch.roll(shifted_x, shifts=(self.shift_size, self.shift_size), dims=(1, 2))
else:
x = shifted_x
if pad_r > 0 or pad_b > 0:
x = x[:, :H, :W, :].contiguous()
x = shortcut + self.drop_path(x)
x = x + self.drop_path(self.mlp(self.norm2(x)))
return x
class PatchMerging(nn.Module):
""" Patch Merging Layer"""
def __init__(self, dim, norm_layer=nn.LayerNorm):
super().__init__()
self.dim = dim
self.reduction = nn.Linear(4 * dim, 2 * dim, bias=False)
self.norm = norm_layer(4 * dim)
def forward(self, x):
B, H, W, C = x.shape
pad_l = pad_t = 0
pad_r = (2 - W % 2) % 2
pad_b = (2 - H % 2) % 2
x = F.pad(x, (0, 0, pad_l, pad_r, pad_t, pad_b))
x0 = x[:, 0::2, 0::2, :]
x1 = x[:, 1::2, 0::2, :]
x2 = x[:, 0::2, 1::2, :]
x3 = x[:, 1::2, 1::2, :]
x = torch.cat([x0, x1, x2, x3], -1)
x = self.norm(x)
x = self.reduction(x)
return x
class PatchExpand(nn.Module):
def __init__(self, dim, norm_layer=nn.LayerNorm):
super().__init__()
self.dim = dim
self.expand = nn.Linear(dim, 2*dim, bias=False)
self.norm = norm_layer(dim // 2)
def forward(self, x):
B, H, W, C = x.shape
x = self.expand(x)
x = x.view(B, H, W, 2, 2, C//2).permute(0,1,3,2,4,5).contiguous().view(B, H*2, W*2, C//2)
x = self.norm(x)
return x
class FinalPatchExpand_X4(nn.Module):
def __init__(self, dim, norm_layer=nn.LayerNorm):
super().__init__()
self.dim = dim
self.expand = nn.Linear(dim, 16*dim, bias=False)
self.norm = norm_layer(dim)
def forward(self, x):
B, H, W, C = x.shape
x = self.expand(x)
x = x.view(B, H, W, 4, 4, C).permute(0,1,3,2,4,5).contiguous().view(B, H*4, W*4, C)
x = self.norm(x)
return x
class BasicLayer(nn.Module):
""" A basic Swin Transformer layer for one stage. """
def __init__(self,
dim,
depth,
num_heads,
window_size=7,
mlp_ratio=4.,
qkv_bias=True,
qk_scale=None,
drop=0.,
attn_drop=0.,
drop_path=0.,
norm_layer=nn.LayerNorm,
downsample=None,
use_checkpoint=False):
super().__init__()
self.window_size = window_size
self.shift_size = window_size // 2
self.depth = depth
self.use_checkpoint = use_checkpoint
self.blocks = nn.ModuleList([
SwinTransformerBlock(
dim=dim,
num_heads=num_heads,
window_size=window_size,
shift_size=0 if (i % 2 == 0) else self.shift_size,
mlp_ratio=mlp_ratio,
qkv_bias=qkv_bias,
qk_scale=qk_scale,
drop=drop,
attn_drop=attn_drop,
drop_path=drop_path[i] if isinstance(drop_path, list) else drop_path,
norm_layer=norm_layer)
for i in range(depth)])
if downsample is not None:
self.downsample = downsample(dim=dim, norm_layer=norm_layer)
else:
self.downsample = None
def forward(self, x):
B, C, H, W = x.shape
# calculate attention mask for SW-MSA
Hp = int(np.ceil(H / self.window_size)) * self.window_size
Wp = int(np.ceil(W / self.window_size)) * self.window_size
img_mask = torch.zeros((1, Hp, Wp, 1), device=x.device)
h_slices = (slice(0, -self.window_size),
slice(-self.window_size, -self.shift_size),
slice(-self.shift_size, None))
w_slices = (slice(0, -self.window_size),
slice(-self.window_size, -self.shift_size),
slice(-self.shift_size, None))
cnt = 0
for h in h_slices:
for w in w_slices:
img_mask[:, h, w, :] = cnt
cnt += 1
mask_windows = window_partition(img_mask, self.window_size)
mask_windows = mask_windows.view(-1, self.window_size * self.window_size)
attn_mask = mask_windows.unsqueeze(1) - mask_windows.unsqueeze(2)
attn_mask = attn_mask.masked_fill(attn_mask != 0, float(-100.0)).masked_fill(attn_mask == 0, float(0.0))
x = x.permute(0, 2, 3, 1) # B H W C
for blk in self.blocks:
x = blk(x, attn_mask)
x = x.permute(0, 3, 1, 2) # B C H W
if self.downsample is not None:
x_down = self.downsample(x.permute(0,2,3,1)).permute(0,3,1,2)
return x, x_down
return x, x
class PatchEmbed(nn.Module):
""" Image to Patch Embedding """
def __init__(self, patch_size=4, in_chans=3, embed_dim=96, norm_layer=None):
super().__init__()
patch_size = to_2tuple(patch_size)
self.patch_size = patch_size
self.in_chans = in_chans
self.embed_dim = embed_dim
self.proj = nn.Conv2d(in_chans, embed_dim, kernel_size=patch_size, stride=patch_size)
if norm_layer is not None:
self.norm = norm_layer(embed_dim)
else:
self.norm = None
def forward(self, x):
_, _, H, W = x.shape
if W % self.patch_size[1] != 0:
x = F.pad(x, (0, self.patch_size[1] - W % self.patch_size[1]))
if H % self.patch_size[0] != 0:
x = F.pad(x, (0, 0, 0, self.patch_size[0] - H % self.patch_size[0]))
x = self.proj(x)
if self.norm is not None:
x = self.norm(x)
return x
class SwinUnet(nn.Module):
def __init__(self, img_size=224, patch_size=4, in_chans=3, num_classes=2,
embed_dim=96, depths=[2, 2, 6, 2], num_heads=[3, 6, 12, 24],
window_size=7, mlp_ratio=4., qkv_bias=True, qk_scale=None,
drop_rate=0., attn_drop_rate=0., drop_path_rate=0.1,
norm_layer=nn.LayerNorm, ape=False, patch_norm=True,
use_checkpoint=False, **kwargs):
super().__init__()
self.num_classes = num_classes
self.num_layers = len(depths)
self.embed_dim = embed_dim
self.ape = ape
self.patch_norm = patch_norm
self.num_features = int(embed_dim * 2 ** (self.num_layers - 1))
self.mlp_ratio = mlp_ratio
# Split image into non-overlapping patches
self.patch_embed = PatchEmbed(
patch_size=patch_size, in_chans=in_chans, embed_dim=embed_dim,
norm_layer=norm_layer if self.patch_norm else None)
# stochastic depth
dpr = [x.item() for x in torch.linspace(0, drop_path_rate, sum(depths))]
# build layers
self.layers = nn.ModuleList()
for i_layer in range(self.num_layers):
layer = BasicLayer(dim=int(embed_dim * 2 ** i_layer),
depth=depths[i_layer],
num_heads=num_heads[i_layer],
window_size=window_size,
mlp_ratio=self.mlp_ratio,
qkv_bias=qkv_bias, qk_scale=qk_scale,
drop=drop_rate, attn_drop=attn_drop_rate,
drop_path=dpr[sum(depths[:i_layer]):sum(depths[:i_layer + 1])],
norm_layer=norm_layer,
downsample=PatchMerging if (i_layer < self.num_layers - 1) else None,
use_checkpoint=use_checkpoint)
self.layers.append(layer)
self.apply(self._init_weights)
def _init_weights(self, m):
if isinstance(m, nn.Linear):
trunc_normal_(m.weight, std=.02)
if isinstance(m, nn.Linear) and m.bias is not None:
nn.init.constant_(m.bias, 0)
elif isinstance(m, nn.LayerNorm):
nn.init.constant_(m.bias, 0)
nn.init.constant_(m.weight, 1.0)
def forward(self, x):
x = self.patch_embed(x)
skips = []
for i, layer in enumerate(self.layers):
x_out, x = layer(x)
if i < self.num_layers -1:
skips.append(x_out)
return x, skips
class DualSwinUnetPlusPlus(SwinUnet):
def __init__(self, img_size=224, patch_size=4, in_chans=1, num_classes=1,
embed_dim=96, depths=[2, 2, 6, 2], depths_decoder=[2, 2, 6, 2],
num_heads=[3, 6, 12, 24], window_size=7, mlp_ratio=4.,
qkv_bias=True, qk_scale=None, drop_rate=0., attn_drop_rate=0.,
drop_path_rate=0.1, norm_layer=nn.LayerNorm, ape=False,
patch_norm=True, use_checkpoint=False, **kwargs):
super().__init__(img_size, patch_size, in_chans, num_classes, embed_dim, depths,
num_heads, window_size, mlp_ratio, qkv_bias, qk_scale, drop_rate,
attn_drop_rate, drop_path_rate, norm_layer, ape, patch_norm, use_checkpoint)
# Encoder
self.encoder = super()
# Bottleneck
self.bottleneck = BasicLayer(dim=self.num_features,
depth=2,
num_heads=num_heads[-1],
window_size=window_size,
mlp_ratio=self.mlp_ratio,
qkv_bias=qkv_bias, qk_scale=qk_scale,
drop=drop_rate, attn_drop=attn_drop_rate,
drop_path=0, norm_layer=norm_layer,
downsample=None, use_checkpoint=use_checkpoint)
# Decoder 1 (Thyroid)
self.decoder1_layers = nn.ModuleList()
for i_layer in range(self.num_layers-1, -1, -1):
layer = self.build_decoder_layer(i_layer, depths_decoder, num_heads, window_size, mlp_ratio, qkv_bias, qk_scale, drop_rate, attn_drop_rate, drop_path_rate, norm_layer, use_checkpoint)
self.decoder1_layers.append(layer)
self.decoder1_up = FinalPatchExpand_X4(embed_dim)
self.decoder1_pred = nn.Conv2d(embed_dim, num_classes, kernel_size=1)
# Decoder 2 (PTMC)
self.decoder2_layers = nn.ModuleList()
for i_layer in range(self.num_layers-1, -1, -1):
layer = self.build_decoder_layer(i_layer, depths_decoder, num_heads, window_size, mlp_ratio, qkv_bias, qk_scale, drop_rate, attn_drop_rate, drop_path_rate, norm_layer, use_checkpoint, is_decoder2=True)
self.decoder2_layers.append(layer)
self.decoder2_up = FinalPatchExpand_X4(embed_dim)
self.decoder2_pred = nn.Conv2d(embed_dim, num_classes, kernel_size=1)
def build_decoder_layer(self, i_layer, depths, num_heads, window_size, mlp_ratio, qkv_bias, qk_scale, drop_rate, attn_drop_rate, drop_path_rate, norm_layer, use_checkpoint, is_decoder2=False):
dim_scale = 2 ** i_layer
up_dim = int(self.embed_dim * dim_scale)
if i_layer > 0:
up_dim = int(self.embed_dim * 2 * dim_scale)
concat_dim_factor = 3 if is_decoder2 and i_layer < self.num_layers-1 else 2
if i_layer == self.num_layers -1:
concat_dim_factor = 2 if is_decoder2 else 1
layer_dim = int(self.embed_dim * dim_scale)
up = PatchExpand(dim=up_dim, norm_layer=norm_layer)
concat_linear = nn.Linear(concat_dim_factor * layer_dim, layer_dim)
layer = BasicLayer(dim=layer_dim,
depth=depths[i_layer],
num_heads=num_heads[i_layer],
window_size=window_size,
mlp_ratio=mlp_ratio,
qkv_bias=qkv_bias, qk_scale=qk_scale,
drop=drop_rate, attn_drop=attn_drop_rate,
drop_path=0, norm_layer=norm_layer,
downsample=None, use_checkpoint=use_checkpoint)
return nn.ModuleDict([
('up', up),
('concat_linear', concat_linear),
('layer', layer)
])
def forward(self, x):
# Encoder
x_enc, skips = self.encoder(x)
# Bottleneck
_, x_bottle = self.bottleneck(x_enc)
# Decoder 1 (Thyroid)
x_dec1 = x_bottle
dec1_skips = []
for i, dec_layer in enumerate(self.decoder1_layers):
skip_idx = self.num_layers - 2 - i
x_dec1 = dec_layer['up'](x_dec1.permute(0,2,3,1)).permute(0,3,1,2)
if skip_idx >= 0:
x_dec1 = torch.cat([x_dec1, skips[skip_idx]], 1)
x_dec1 = dec_layer['concat_linear'](x_dec1.permute(0,2,3,1)).permute(0,3,1,2)
_, x_dec1 = dec_layer['layer'](x_dec1)
if skip_idx >= 0:
dec1_skips.append(x_dec1)
thyroid_out = self.decoder1_up(x_dec1.permute(0,2,3,1)).permute(0,3,1,2)
thyroid_out = self.decoder1_pred(thyroid_out)
# Decoder 2 (PTMC)
x_dec2 = x_bottle
for i, dec_layer in enumerate(self.decoder2_layers):
skip_idx = self.num_layers - 2 - i
x_dec2 = dec_layer['up'](x_dec2.permute(0,2,3,1)).permute(0,3,1,2)
if skip_idx >= 0:
# Concatenate encoder skip, and decoder1 skip
x_dec2 = torch.cat([x_dec2, skips[skip_idx], dec1_skips[i]], 1)
x_dec2 = dec_layer['concat_linear'](x_dec2.permute(0,2,3,1)).permute(0,3,1,2)
_, x_dec2 = dec_layer['layer'](x_dec2)
ptmc_out = self.decoder2_up(x_dec2.permute(0,2,3,1)).permute(0,3,1,2)
ptmc_out = self.decoder2_pred(ptmc_out)
return torch.sigmoid(thyroid_out), torch.sigmoid(ptmc_out)
# Loss Function as described in the paper
class DiceBCELoss(nn.Module):
def __init__(self, weight=None, size_average=True):
super(DiceBCELoss, self).__init__()
def forward(self, inputs, targets, smooth=1):
#comment out if your model contains a sigmoid or equivalent activation layer
# inputs = F.sigmoid(inputs)
#flatten label and prediction tensors
inputs = inputs.view(-1)
targets = targets.view(-1)
intersection = (inputs * targets).sum()
dice_loss = 1 - (2.*intersection + smooth)/(inputs.sum() + targets.sum() + smooth)
BCE = F.binary_cross_entropy(inputs, targets, reduction='mean')
Dice_BCE = BCE + dice_loss
return Dice_BCE
class DualSwinUnetPlusPlusLoss(nn.Module):
def __init__(self, alpha=0.5, beta=0.5):
super().__init__()
self.alpha = alpha
self.beta = beta
self.dice_bce_loss = DiceBCELoss()
def forward(self, pred_thyroid, true_thyroid, pred_ptmc, true_ptmc):
loss_thyroid = self.alpha * F.binary_cross_entropy(pred_thyroid, true_thyroid) + \
(1 - self.alpha) * (1 - self.dice_coeff(pred_thyroid, true_thyroid))
loss_ptmc = self.beta * F.binary_cross_entropy(pred_ptmc, true_ptmc) + \
(1 - self.beta) * (1 - self.dice_coeff(pred_ptmc, true_ptmc))
total_loss = (loss_thyroid + loss_ptmc) / 2
return total_loss
def dice_coeff(self, pred, target, smooth=1.):
pred = pred.contiguous().view(pred.shape[0], -1)
target = target.contiguous().view(target.shape[0], -1)
intersection = torch.sum(pred * target, dim=1)
dice = (2. * intersection + smooth) / (torch.sum(pred, dim=1) + torch.sum(target, dim=1) + smooth)
return dice.mean()
if __name__ == '__main__':
# Example Usage
# Hyperparameters from the paper (Table 6)
img_size = 224
patch_size = 4
in_chans = 1 # Grayscale ultrasound
num_classes = 1 # Binary segmentation for each decoder
model = DualSwinUnetPlusPlus(
img_size=img_size,
patch_size=patch_size,
in_chans=in_chans,
num_classes=num_classes,
embed_dim=96,
depths=[2, 2, 6, 2],
depths_decoder=[2, 2, 6, 2],
num_heads=[3, 6, 12, 24],
window_size=7,
mlp_ratio=4.,
qkv_bias=True,
qk_scale=None,
drop_rate=0.0,
attn_drop_rate=0.0,
drop_path_rate=0.1,
patch_norm=True
)
# Dummy input
dummy_input = torch.randn(2, in_chans, img_size, img_size) # Batch size of 2
# Forward pass
pred_thyroid, pred_ptmc = model(dummy_input)
print("Model instantiated successfully.")
print(f"Input shape: {dummy_input.shape}")
print(f"Predicted Thyroid Mask shape: {pred_thyroid.shape}")
print(f"Predicted PTMC Mask shape: {pred_ptmc.shape}")
# Example loss calculation
true_thyroid_mask = torch.randint(0, 2, (2, 1, img_size, img_size)).float()
true_ptmc_mask = torch.randint(0, 2, (2, 1, img_size, img_size)).float()
criterion = DualSwinUnetPlusPlusLoss(alpha=0.5, beta=0.5)
loss = criterion(pred_thyroid, true_thyroid_mask, pred_ptmc, true_ptmc_mask)
print(f"Calculated Loss: {loss.item()}")
# --- Placeholder for Training Loop ---
# optimizer = torch.optim.AdamW(model.parameters(), lr=1e-4, weight_decay=0.05)
#
# for epoch in range(num_epochs):
# for images, thyroid_masks, ptmc_masks in train_loader:
# # Move data to device
# images = images.to(device)
# thyroid_masks = thyroid_masks.to(device)
# ptmc_masks = ptmc_masks.to(device)
#
# # Forward pass
# pred_thyroid, pred_ptmc = model(images)
#
# # Calculate loss
# loss = criterion(pred_thyroid, thyroid_masks, pred_ptmc, ptmc_masks)
#
# # Backward and optimize
# optimizer.zero_grad()
# loss.backward()
# optimizer.step()
#
# print(f"Epoch [{epoch+1}/{num_epochs}], Loss: {loss.item():.4f}")

Pingback: 7 Revolutionary Breakthroughs in Cardiac Motion Analysis: How a New AI Model Outperforms Old Methods (And Why It Matters) - aitrendblend.com
Pingback: 7 Revolutionary Breakthroughs in NPH Diagnosis: the Future of AI-Powered Brain Scans - aitrendblend.com
Pingback: 7 Revolutionary Breakthroughs in Bacteria Detection: How EmiNet Outperforms Old Methods - aitrendblend.com
Pingback: 3 Revolutionary FCS-MPDTC Breakthroughs That Slash Energy Waste in Linear Motors - aitrendblend.com
Pingback: 7 Revolutionary Breakthroughs in Knowledge Distillation: Why Swapped Logit Distillation Outperforms Old Methods - aitrendblend.com
Your article helped me a lot, is there any more related content? Thanks!