Introduction: The Critical Need for Intelligent, Privacy-Preserving Skin Cancer Diagnosis
Skin cancer remains one of the most pervasive and life-threatening health conditions globally, with over 5 million new cases reported annually in the United States alone. Among the various types, malignant melanoma stands out as particularly alarming—accounting for approximately 4% of global cancer-related deaths and roughly 10,000 deaths annually in the U.S. Despite advances in medical imaging, traditional diagnostic methods, including visual dermoscopic examination and biopsies, achieve only approximately 60% accuracy, leaving significant room for improvement.
The convergence of artificial intelligence and healthcare has opened unprecedented opportunities for enhancing diagnostic precision. However, deploying AI in medical settings introduces a fundamental paradox: how do we train sophisticated diagnostic models that require vast amounts of data while rigorously protecting sensitive patient information? Centralized machine learning approaches, though effective, demand aggregating patient data on single servers—creating unacceptable privacy risks and regulatory complications in an era of stringent data protection laws.
Enter Multimodal Adaptive Federated Learning Network (MAFL-Net), a groundbreaking framework that elegantly solves this dilemma. By synergistically integrating multimodal dermoscopic imaging with structured clinical metadata within a privacy-preserving federated learning environment, MAFL-Net achieves diagnostic accuracies of 99.65% on HAM10000, 98.87% on ISIC 2019, and 95.46% on ISIC 2024 datasets—outperforming state-of-the-art methods while ensuring patient data never leaves local institutions.
This article explores how MAFL-Net represents a paradigm shift in dermatological AI, combining computer vision, natural language processing, and distributed machine learning to deliver superior diagnostic capabilities without compromising patient privacy.
Understanding the Limitations of Current Skin Cancer Detection Approaches
The Data Privacy Conundrum in Medical AI
Traditional AI development in healthcare follows a centralized paradigm: hospitals and clinics share patient data with a central server where models are trained. While this approach maximizes data availability, it creates critical vulnerabilities:
- Privacy breaches expose sensitive health records to unauthorized access
- Regulatory non-compliance with HIPAA, GDPR, and other data protection frameworks
- Institutional resistance to data sharing due to liability concerns
- Patient trust erosion when personal health information is transmitted across networks
Key Takeaway: The medical AI community faces an urgent need for training methodologies that preserve diagnostic accuracy while eliminating centralized data aggregation.
The Unimodal Limitation
Most existing skin cancer detection systems rely exclusively on either:
- Dermoscopic images alone, missing crucial contextual patient information
- Clinical metadata alone, ignoring visual lesion characteristics
This unimodal approach fundamentally limits diagnostic accuracy because dermatological assessment inherently combines visual pattern recognition with patient history, demographics, and anatomical context.
Class Imbalance and Dataset Bias
Publicly available dermatological datasets exhibit severe class imbalances. For instance, the ISIC 2024 dataset contains 400,666 benign samples versus only 393 malignant cases—a ratio exceeding 1000:1. Without sophisticated handling, models trained on such data become biased toward majority classes, potentially missing life-threatening malignancies.
MAFL-Net: Architecture and Technical Innovation
Core Concept: Late-Fusion Multimodal Learning
MAFL-Net employs a late-fusion strategy that processes dermoscopic images and clinical metadata through separate, optimized pathways before combining high-level representations. This approach offers distinct advantages over early fusion:
| Fusion Strategy | Advantages | Disadvantages |
|---|---|---|
| Early Fusion | Simple implementation, unified feature space | Modality dominance, noise amplification, loss of modality-specific patterns |
| Late Fusion (MAFL-Net) | Preserves modality-specific features, reduces cross-modal interference, enables specialized processing for each data type | Increased architectural complexity, higher computational requirements |
Bold Insight: Late fusion allows each modality to develop its optimal representation before combination, preventing “modality dominance” where stronger signals (typically visual) suppress clinically relevant metadata patterns.
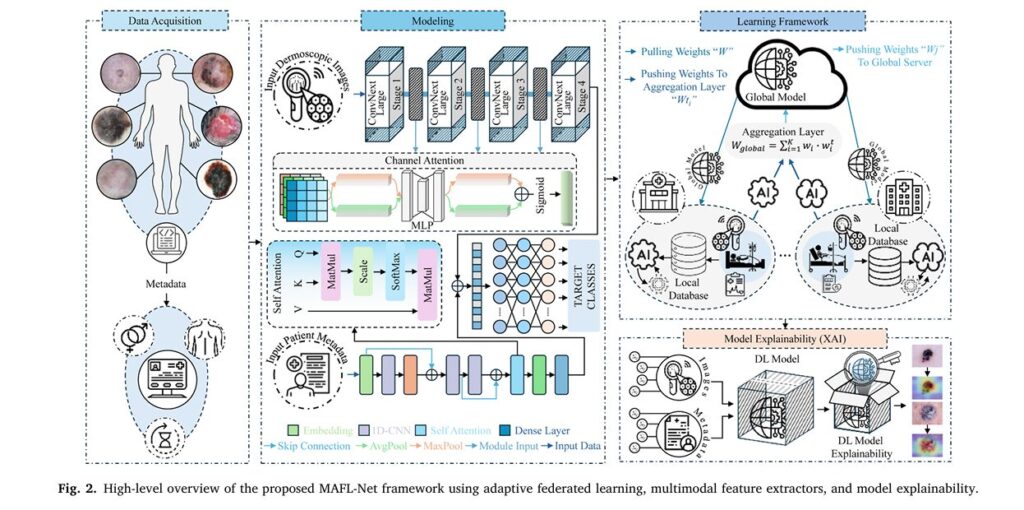
Visual Feature Extraction: ConvNeXt-Large with Channel Attention
The image processing pipeline leverages ConvNeXt-Large, a modern convolutional architecture that incorporates transformer-inspired design principles while maintaining convolutional efficiency. The architecture processes dermoscopic images through four hierarchical stages:
\[ x_i’ = \sigma_i + \epsilon \frac{x_i – \mu_i}{ } \]This layer normalization equation ensures consistent feature scaling across batches, enhancing training stability. Each stage employs:
- 7×7 depthwise separable convolutions for efficient spatial feature extraction
- Inverted bottleneck blocks that expand channel dimensions to increase representational capacity
- Layer normalization replacing batch normalization for improved stability
- Channel Attention Modules (CAM) after each stage to emphasize diagnostically relevant regions
The Channel Attention Mechanism mathematically operates as:
\[ \mathrm{CAM}(X) = X \cdot \mathrm{Sigmoid} \Big( W_2 \big( \mathrm{ReLU}( W_1 \cdot \mathrm{AvgPool}(X) ) \big) + W_2 \big( \mathrm{ReLU}( W_1 \cdot \mathrm{MaxPool}(X) ) \big) \Big) \]Where:
- W1,W2 represent 1×1 convolutional layers for dimensionality reduction and expansion
- Average pooling captures global statistical distributions
- Max pooling preserves the most salient spatial features
- The sigmoid activation generates attention weights between 0 and 1
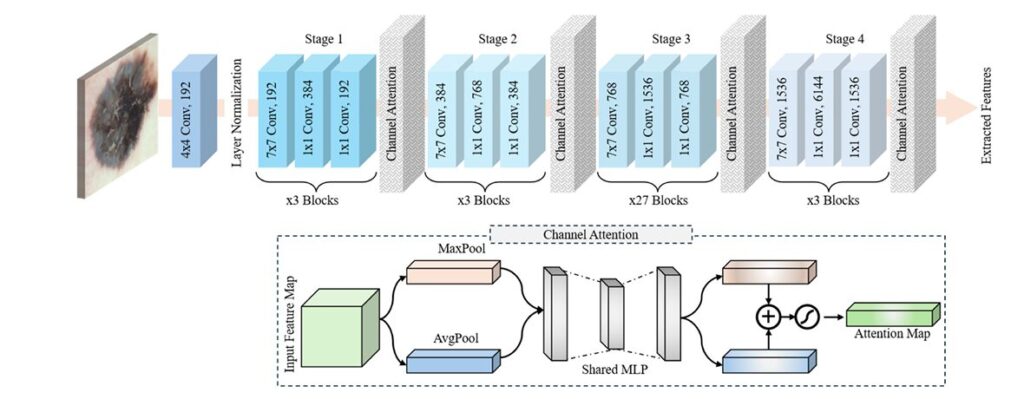
Clinical Metadata Processing: 1D-CNN with Self-Attention
The metadata pathway transforms structured clinical attributes (age, gender, lesion location) into meaningful representations through an innovative sentence structuring approach. Rather than using raw categorical values, MAFL-Net constructs natural language sentences:
“The patient, identified as [gender], aged [age], has skin cancer located in the [localization] region.”
This semantic structuring enables richer contextual understanding. The text processing pipeline includes:
- Tokenization and embedding (64-dimensional vectors)
- 1D-CNN with skip connections for local dependency capture and gradient flow improvement
- Self-Attention Mechanism (SAM) for long-range dependency modeling
The Self-Attention Mechanism computes attention scores through scaled dot-product attention:
\[ \mathrm{Attention}(Q, K, V) = \mathrm{SoftMax}\!\left( \frac{Q K^{\mathrm{T}}}{\sqrt{d_k}} \right) V \]Where Q (queries), K (keys), and V (values) are derived from the input sequence through learned projection matrices WQ , WK , and WV .
Joint Feature Fusion and Classification
The multimodal fusion combines:
- Visual features: 512-dimensional vector from ConvNeXt-Large
- Textual features: 64-dimensional vector from metadata processing
This 576-dimensional joint representation passes through fully connected layers (576 → 64 → 32) before final classification, enabling the model to learn complex cross-modal interactions.
Adaptive Federated Averaging (AdaFedAvg): Privacy-Preserving Collaborative Learning
The Federated Learning Paradigm
Federated Learning (FL), introduced by Google in 2016, enables decentralized model training across multiple institutions without sharing raw data. In MAFL-Net’s FL environment:
- Each hospital trains a local model on its private dataset
- Only model parameters (weights and gradients) are transmitted to a central server
- The server aggregates updates and distributes the improved global model
- Patient data never leaves local premises
Mathematical Formulation of Distributed Learning
The global training set Dtrain is partitioned across K clients using a Dirichlet distribution with concentration parameter α=0.1 to simulate realistic non-IID (non-Independent and Identically Distributed) data distributions:
\[ D_{\text{train}} = \bigcup_{k=1}^{K} D_k, \qquad \text{where } D_i \cap D_j = \varnothing \;\; \text{for } i \ne j. \]Each client k minimizes its local loss function:
\[ F_k(w) = \frac{1}{\left| D_k \right|} \sum_{i \in D_k} \ell\!\left(w; x_i, y_i\right) \]Using Categorical Cross-Entropy Loss:
\[ \text{CCE} = – \sum_{i=1}^{C} y_i \cdot \log(y_i’) \]Where c represents the number of classes, yi is the ground truth, and yi′ is the predicted probability for class i .
Adaptive Aggregation: Weighting by Validation Performance
Traditional FedAvg assigns equal weights to all client updates, which proves suboptimal under heterogeneous data distributions. AdaFedAvg introduces performance-based adaptive weighting:
\[ W_{t+1} = \sum_{i=1}^{N} a_{i}^{t} \, w_{i}^{t} \]With adaptive weights computed as:
\[ a_{i t} = \sum_{j=1}^{N} A_{j t} \, A_{i t} \]Where Ait represents the validation accuracy of client i at communication round t .
Strategic Advantage: This validation-driven weighting ensures that clients with higher-quality data or better local optimization contribute more significantly to the global model, accelerating convergence and improving final performance.
Experimental Validation: Benchmarking Against State-of-the-Art
Dataset Characteristics and Preprocessing
MAFL-Net was rigorously evaluated on three challenging benchmarks:
| Dataset | Year | Classes | Total Samples | Key Characteristics |
|---|---|---|---|---|
| HAM10000 | 2018 | 7 (NV, MEL, BKL, BCC, AKIEC, VASC, DF) | 10,015 | Highly imbalanced, diverse lesion types |
| ISIC 2019 | 2019 | 8 (+ SCC) | 25,331 | Large-scale, includes clinical metadata |
| ISIC 2024 | 2024 | 2 (Benign/Malignant) | 401,059 | Extreme class imbalance (1000:1 ratio), 3D Total Body Photography |
Preprocessing innovations include:
- Dataset-specific resampling: Upsampling minority classes in HAM10000/ISIC 2019; strategic undersampling/upsampling (10:1 ratio) in ISIC 2024
- Extensive augmentation: Vertical/horizontal flipping, random rotations, resizing, cropping, and enhancement transformations
- Metadata augmentation tracking: Each image transformation is annotated in corresponding metadata to maintain consistency
Quantitative Performance Analysis
Table: Comparative Performance on Benchmark Datasets
| Method | Type | HAM10000 Acc | ISIC 2019 Acc | Key Limitations |
|---|---|---|---|---|
| Houssein et al. (2024) | Transfer Learning | 98.50% | 97.11% | Unimodal, centralized |
| Chanda et al. (2024) | Ensemble | 99.50% | — | High computational cost, no privacy preservation |
| Naeem et al. (2022) | Deep Learning | — | 96.91% | Centralized training, single modality |
| MAFL-Net (Proposed) | Multimodal FL | 99.65% | 98.87% | Privacy-preserving, multimodal, distributed |
Critical Insight: MAFL-Net achieves 99.65% accuracy on HAM10000 and 98.87% on ISIC 2019, surpassing even sophisticated ensemble methods while maintaining strict privacy guarantees. On the challenging ISIC 2024 dataset with extreme imbalance, it achieves 95.46% accuracy—remarkable given the 1000:1 class ratio.
Federated Learning Algorithm Comparison
Under severe non-IID conditions (α=0.1 ), AdaFedAvg demonstrates superior stability and convergence:
| Algorithm | HAM10000 Val Acc | ISIC 2019 Val Acc | ISIC 2024 Val Acc | Key Feature |
|---|---|---|---|---|
| FedAvg | 94.83% | 96.60% | 93.82% | Uniform weighting |
| FedProx | 95.50% | 90.03% | 93.61% | Proximal term for heterogeneity |
| FedFuse | 93.04% | 97.10% | 93.34% | Adaptive fusion |
| AdaFedAvg | 99.65% | 98.87% | 95.46% | Validation-accuracy weighting |
Qualitative Assessment: Clinical Alignment
Beyond accuracy metrics, MAFL-Net demonstrates strong clinical interpretability through Grad-CAM visualizations. The model achieves:
- Mean IoU of 89.53% on HAM10000 between attention maps and dermatologist-annotated regions
- Mean IoU of 85.25% on ISIC 2019
- Mean IoU of 86.26% on ISIC 2024
Clinical Significance: High Intersection over Union (IoU) scores indicate that MAFL-Net focuses on clinically relevant lesion regions—such as asymmetric borders, color variations, and irregular structures—aligning with dermatologist assessment patterns based on the ABCD rule (Asymmetry, Border irregularity, Color, Diameter).
Technical Innovations and Architectural Advantages
Why ConvNeXt-Large Outperforms Alternatives
Ablation studies comparing visual backbones reveal:
| Backbone | HAM10000 Acc | GFLOPs | Memory (MB) | Latency (ms) |
|---|---|---|---|---|
| MobileNet-V3 | 99.12% | 0.26 | 187.49 | 6.35 |
| EfficientNet-B0 | 97.67% | 0.45 | 261.18 | 6.42 |
| ConvNeXt-Small | 99.32% | 8.72 | 490.78 | 8.84 |
| ConvNeXt-Large (MAFL-Net) | 99.65% | 34.40 | 1212.64 | 10.35 |
While lightweight models offer speed advantages, ConvNeXt-Large with Channel Attention provides the optimal accuracy-complexity trade-off for diagnostic applications where precision is paramount.
Metadata Processing: Custom 1D-CNN vs. Pretrained Language Models
Surprisingly, the proposed 1D-CNN with skip connections and Self-Attention outperforms heavyweight pretrained language models:
| Text Encoder | HAM10000 Acc | Parameters (M) | Inference Time |
|---|---|---|---|
| BioBERT | 98.94% | 110M | 18.72ms |
| ClinicalBERT | 99.05% | 110M | 18.72ms |
| BlueBERT | 98.93% | 110M | 18.72ms |
| 1D-CNN + Skip + SAM (MAFL-Net) | 99.65% | 0.2M | 10.35ms |
Key Finding: For structured clinical metadata (age, gender, location), task-specific lightweight architectures outperform general-purpose language models that are pretrained on unstructured biomedical text, while requiring 550× fewer parameters.
Real-World Implications and Future Directions
Clinical Deployment Considerations
MAFL-Net’s architecture addresses critical requirements for real-world dermatological AI:
✅ Privacy Compliance: Federated learning eliminates HIPAA/GDPR concerns associated with data centralization
✅ Institutional Collaboration: Enables multi-hospital studies without data sharing agreements
✅ Diagnostic Transparency: Attention maps provide interpretable visual explanations for clinicians
✅ Scalability: Adaptive aggregation accommodates varying data quality across institutions
✅ Robustness: Validated across diverse datasets with different imaging protocols
Current Limitations and Research Opportunities
Computational Intensity: The ConvNeXt-Large backbone, while effective, requires significant memory (1.2GB) and processing power. Future work should explore:
- Knowledge distillation to compress models for edge deployment
- Neural architecture search for dermatology-specific efficient networks
- Quantization and pruning techniques for mobile dermoscopy devices
Security in Federated Settings: FL is vulnerable to:
- Model poisoning attacks from malicious clients submitting corrupted updates
- Gradient leakage potentially reconstructing sensitive information from shared gradients
- Byzantine failures where unreliable clients degrade global model performance
Next-Generation Solutions include secure aggregation protocols, differential privacy mechanisms, and Byzantine-robust optimization strategies.
Dataset Diversity: Current validation relies on public datasets with standardized acquisition conditions. Future studies must validate on:
- Multi-center clinical data from diverse geographic regions
- Various dermoscopy device manufacturers
- Different skin types and ethnic populations
- Real-world lighting and environmental conditions
Conclusion: A New Era for Privacy-Preserving Medical AI
MAFL-Net represents a paradigm shift in dermatological diagnostics, demonstrating that multimodal federated learning can simultaneously achieve superior accuracy and rigorous privacy protection. By intelligently fusing visual dermoscopic patterns with structured clinical metadata through adaptive distributed optimization, the framework achieves near-perfect diagnostic performance while ensuring sensitive patient data remains under institutional control.
The implications extend far beyond skin cancer detection. The architectural principles—late multimodal fusion, attention-enhanced feature extraction, and performance-weighted federated aggregation—provide a template for privacy-preserving AI across medical specialties, from radiology to pathology to genomics.
For healthcare institutions, MAFL-Net offers a pathway to collaborative AI development without regulatory risk. For patients, it promises more accurate diagnoses without compromising personal health information. For researchers, it establishes new benchmarks for multimodal medical AI architecture.
Call to Action: Join the Privacy-Preserving Medical AI Revolution
The future of healthcare AI depends on our ability to balance diagnostic excellence with ethical data stewardship. Whether you’re a dermatologist seeking to implement AI in your practice, a machine learning engineer developing medical AI systems, a healthcare administrator evaluating privacy-preserving technologies, or a researcher advancing federated learning methodologies—your engagement is crucial.
Here’s how you can contribute today:
🔬 Clinicians: Evaluate MAFL-Net’s attention visualizations against your diagnostic workflow—does the model focus on regions matching your clinical assessment?
💻 Technologists: Implement the AdaFedAvg algorithm in your federated learning projects and share performance benchmarks on diverse medical datasets
🏥 Healthcare Leaders: Pilot federated learning collaborations with partner institutions to build robust, generalizable diagnostic models
📊 Researchers: Address the identified limitations—develop lightweight architectures for resource-constrained settings and security protocols for adversarial FL environments
Share your thoughts in the comments: How do you see federated learning transforming medical AI in your field? What privacy challenges has your institution faced in AI adoption?
Subscribe to our newsletter for the latest advances in privacy-preserving medical AI, multimodal machine learning, and healthcare technology innovation.
Below is a comprehensive, end-to-end implementation of the MAFL-Net framework based on the research paper. This will include all components: data preprocessing, multimodal feature extraction, federated learning with AdaFedAvg, and training/evaluation pipelines.
"""
requirements.txt for MAFL-Net
torch>=2.0.0
torchvision>=0.15.0
timm>=0.9.0 # For ConvNeXt models
transformers>=4.30.0 # For BERT-based alternatives (optional)
pandas>=1.5.0
numpy>=1.24.0
Pillow>=9.5.0
scikit-learn>=1.2.0
matplotlib>=3.7.0
seaborn>=0.12.0
tqdm>=4.65.0
tensorboard>=2.13.0
"""
"""
data_preparation.py - Helper script for dataset preparation
This script helps prepare HAM10000, ISIC 2019, and ISIC 2024 datasets
for use with MAFL-Net.
"""
import os
import pandas as pd
import numpy as np
from pathlib import Path
import shutil
class DatasetPreparer:
"""
Prepares and validates skin lesion datasets for MAFL-Net training.
"""
HAM10000_CLASSES = ['nv', 'mel', 'bkl', 'bcc', 'akiec', 'vasc', 'df']
ISIC2019_CLASSES = ['nv', 'mel', 'bkl', 'bcc', 'akiec', 'vasc', 'df', 'scc']
ISIC2024_CLASSES = ['benign', 'malignant']
def __init__(self, root_dir: str):
self.root_dir = Path(root_dir)
def prepare_ham10000(self, metadata_path: str, images_dir: str):
"""
Prepare HAM10000 dataset with proper metadata formatting.
"""
df = pd.read_csv(metadata_path)
# Standardize column names
column_mapping = {
'image_id': 'image_id',
'dx': 'label',
'age': 'age',
'sex': 'sex',
'localization': 'localization'
}
df = df.rename(columns=column_mapping)
# Ensure consistent label naming
df['label'] = df['label'].str.lower()
# Validate classes
unique_labels = df['label'].unique()
print(f"HAM10000 classes found: {unique_labels}")
# Handle missing values
df['age'] = df['age'].fillna(df['age'].median())
df['sex'] = df['sex'].fillna('unknown')
df['localization'] = df['localization'].fillna('unknown')
# Save processed metadata
output_path = self.root_dir / 'HAM10000_processed.csv'
df.to_csv(output_path, index=False)
print(f"Saved processed metadata to {output_path}")
return df
def prepare_isic2019(self, metadata_path: str):
"""
Prepare ISIC 2019 dataset.
"""
df = pd.read_csv(metadata_path)
# ISIC 2019 specific processing
df['label'] = df['diagnosis'].str.lower()
# Standardize metadata
df['image_id'] = df['image_name']
df['sex'] = df['sex'].fillna('unknown')
df['age'] = df['age_approx'].fillna(50)
df['localization'] = df['anatom_site_general_challenge'].fillna('unknown')
output_path = self.root_dir / 'ISIC2019_processed.csv'
df.to_csv(output_path, index=False)
return df
def prepare_isic2024(self, metadata_path: str):
"""
Prepare ISIC 2024 dataset with binary classification.
"""
df = pd.read_csv(metadata_path)
# Binary classification: benign vs malignant
df['label'] = df['malignant'].map({0: 'benign', 1: 'malignant'})
# Extract relevant columns
df['image_id'] = df['isic_id']
df['age'] = df['age_approx']
df['sex'] = df['sex'].fillna('unknown')
df['localization'] = df['anatom_site_general'].fillna('unknown')
output_path = self.root_dir / 'ISIC2024_processed.csv'
df.to_csv(output_path, index=False)
return df
def verify_dataset(self, metadata_df: pd.DataFrame, images_dir: str):
"""
Verify that all images referenced in metadata exist.
"""
missing = []
found = 0
for img_id in metadata_df['image_id']:
# Check multiple extensions
for ext in ['.jpg', '.jpeg', '.png']:
if (Path(images_dir) / f"{img_id}{ext}").exists():
found += 1
break
else:
missing.append(img_id)
print(f"Images found: {found}/{len(metadata_df)}")
if missing:
print(f"Missing images: {len(missing)}")
print(f"First 5 missing: {missing[:5]}")
return len(missing) == 0
"""
inference.py - Inference script for trained MAFL-Net models
"""
import torch
import torch.nn.functional as F
from PIL import Image
import json
class MAFLNetInference:
"""
Inference pipeline for deployed MAFL-Net models.
"""
def __init__(self, model_path: str, config_path: str = None):
self.device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
# Load model
checkpoint = torch.load(model_path, map_location=self.device)
from maflnet import MAFLNet, Config # Import from main file
self.config = Config()
self.model = MAFLNet(num_classes=checkpoint.get('num_classes', 7))
self.model.load_state_dict(checkpoint['model_state_dict'])
self.model.eval().to(self.device)
self.metadata_processor = MetadataProcessor()
# Load class names if available
self.class_names = checkpoint.get('class_names',
['AKIEC', 'BCC', 'BKL', 'DF', 'MEL', 'NV', 'VASC'])
def predict(self, image_path: str, age: float, gender: str, localization: str):
"""
Make prediction on single sample.
Args:
image_path: Path to dermoscopic image
age: Patient age
gender: 'male', 'female', or 'unknown'
localization: Body location of lesion
Returns:
Dictionary with prediction results
"""
# Load and preprocess image
image = Image.open(image_path).convert('RGB')
transform = DataAugmentation.get_val_transforms(self.config.IMAGE_SIZE)
image_tensor = transform(image).unsqueeze(0).to(self.device)
# Process metadata
metadata_sentence = self.metadata_processor.structure_metadata(
age, gender, localization
)
metadata_tokens = self.metadata_processor.tokenize_metadata(metadata_sentence)
metadata_tensor = metadata_tokens.unsqueeze(0).to(self.device)
# Inference
with torch.no_grad():
logits = self.model(image_tensor, metadata_tensor)
probabilities = F.softmax(logits, dim=1)
# Get prediction
confidence, predicted_class = probabilities.max(1)
# Top-3 predictions
top3_prob, top3_idx = probabilities.topk(3, dim=1)
results = {
'predicted_class': self.class_names[predicted_class.item()],
'confidence': confidence.item() * 100,
'all_probabilities': {
cls: prob.item() * 100
for cls, prob in zip(self.class_names, probabilities[0])
},
'top3_predictions': [
{'class': self.class_names[idx], 'probability': prob.item() * 100}
for idx, prob in zip(top3_idx[0], top3_prob[0])
],
'metadata_processed': metadata_sentence
}
return results
def predict_batch(self, samples: list):
"""
Batch prediction for multiple samples.
"""
results = []
for sample in samples:
result = self.predict(
sample['image_path'],
sample['age'],
sample['gender'],
sample['localization']
)
results.append(result)
return results
# Example usage
if __name__ == "__main__":
# Initialize inference
# inferencer = MAFLNetInference('./checkpoints/maflnet_best.pth')
# Single prediction
# result = inferencer.predict(
# image_path='./sample_lesion.jpg',
# age=55,
# gender='male',
# localization='back'
# )
# print(json.dumps(result, indent=2))
passRelated posts, You May like to read
- 7 Shocking Truths About Knowledge Distillation: The Good, The Bad, and The Breakthrough (SAKD)
- TransXV2S-Net: Revolutionary AI Architecture Achieves 95.26% Accuracy in Skin Cancer Detection
- TimeDistill: Revolutionizing Time Series Forecasting with Cross-Architecture Knowledge Distillation
- HiPerformer: A New Benchmark in Medical Image Segmentation with Modular Hierarchical Fusion
- GeoSAM2 3D Part Segmentation — Prompt-Controllable, Geometry-Aware Masks for Precision 3D Editing
- DGRM: How Advanced AI is Learning to Detect Machine-Generated Text Across Different Domains
- A Knowledge Distillation-Based Approach to Enhance Transparency of Classifier Models
- Towards Trustworthy Breast Tumor Segmentation in Ultrasound Using AI Uncertainty
- Discrete Migratory Bird Optimizer with Deep Transfer Learning for Multi-Retinal Disease Detection
- How AI Combines Medical Images and Patient Data to Detect Skin Cancer More Accurately: A Deep Dive into Multimodal Deep Learning