Early Detection, Smarter AI: How PSO-Optimized Fractional Order CNNs Are Transforming Breast Cancer Diagnosis
Every year, millions of women face the daunting challenge of a breast cancer diagnosis. Despite advances in medical imaging, traditional mammography still struggles with high false-positive and false-negative rates, especially in patients with dense breast tissue. These limitations can lead to unnecessary biopsies, delayed treatment, and increased patient anxiety.
But what if artificial intelligence could see what the human eye—and even conventional AI—might miss?
A groundbreaking new study introduces a powerful solution: PSO-optimized Fractional Order Convolutional Neural Networks (Frac-CNNs). This innovative approach combines advanced image filtering, fractional calculus, and evolutionary optimization to achieve unprecedented accuracy in breast cancer detection—99.35% accuracy, 99% sensitivity, and 98.2% specificity.
In this article, we’ll break down how this cutting-edge AI system works, why it outperforms existing models, and what it means for the future of medical diagnostics.
What Are PSO-Optimized Fractional Order CNNs?
At the heart of this breakthrough is the PSO-optimized Fractional Order CNN (Frac-CNN)—a deep learning architecture that redefines how neural networks process medical images.
Let’s unpack that:
- Convolutional Neural Networks (CNNs) are the gold standard for image analysis, capable of identifying patterns in pixels.
- Fractional Order CNNs (Frac-CNNs) go a step further by incorporating fractional calculus, allowing the network to model complex, non-linear patterns in mammograms with greater precision.
- Particle Swarm Optimization (PSO) fine-tunes the model’s hyperparameters (like learning rate and dropout) to ensure peak performance.
Together, these components create a system that doesn’t just classify images—it understands them at a deeper level.
Why Traditional CNNs Fall Short in Breast Cancer Detection
Standard CNNs use integer-order derivatives in their activation functions (like ReLU). While effective, they struggle with:
- Irregular tumor boundaries
- Subtle texture changes
- Overlapping tissue structures
These are common in dense breasts, where cancer can hide in plain sight.
Fractional calculus addresses this by introducing a fractional-order activation function, which provides a smoother, more flexible response to input data.
The Fractional ReLU (Frc-ReLU) layer is defined as:
\[ R(x) = \begin{cases} x^{\alpha_0}, & \text{if } x \geq 0 \\ x, & \text{if } x < 0 \end{cases} \]Where:
- x is the input neuron value
- α is the fractional order parameter (optimized via PSO)
This allows the model to capture fine-grained variations in pixel intensity, making it far more sensitive to early-stage malignancies.
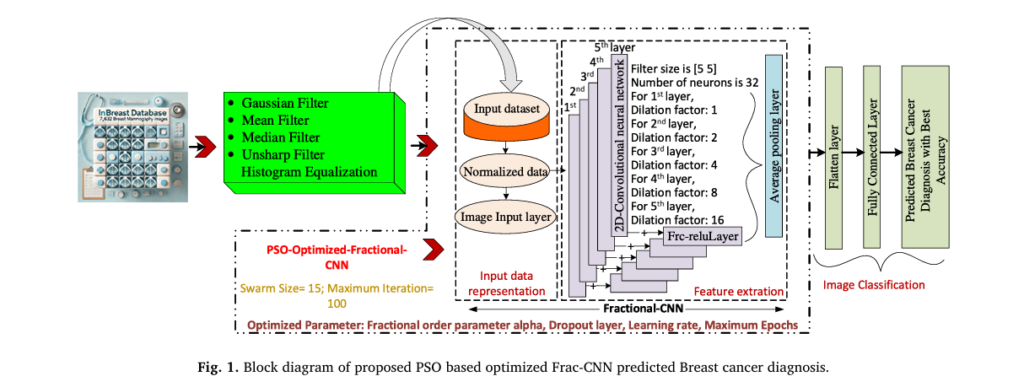
How the System Works: A Step-by-Step Breakdown
The proposed methodology follows a robust pipeline designed to maximize diagnostic accuracy.
1. Data Acquisition and Augmentation
The model was trained on 7,632 mammography images from the INbreast database—a high-quality, annotated dataset widely used in medical AI research.
To combat data scarcity, the team applied data augmentation techniques:
- Multi-angle rotation (30° to 330°)
- Horizontal and vertical flipping
This expanded dataset diversity without compromising privacy.
2. Advanced Image Filtering for Noise Reduction
Before feeding images into the Frac-CNN, they undergo enhanced preprocessing to improve clarity and reduce noise.
The researchers tested four filter combinations and found that Gaussian + Mean + Histogram Equalization delivered the best balance of noise reduction and feature enhancement.
| FILTER | PURPOSE |
|---|---|
| Gaussian Filter | Reduces Gaussian noise (common in low-dose imaging) |
| Mean Filter | Smooths image by averaging pixel values |
| Histogram Equalization | Enhances contrast for better feature visibility |
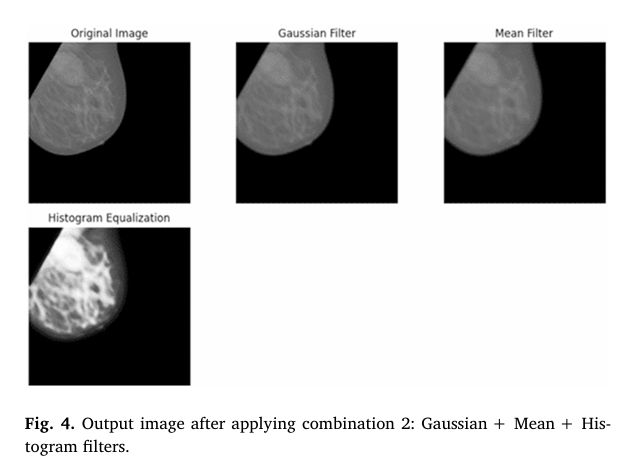
This preprocessing step is crucial. As the study notes, adaptive mean filtering achieves 95% noise reduction, outperforming traditional Gaussian filters (85%).
3. Frac-CNN Architecture: Capturing Complex Patterns
The Frac-CNN architecture includes:
- Input Layer: 150×150×3 RGB images
- Convolutional Layers: 5 layers with 32 filters each, using dilation factors (1, 2, 4, 8, 16)
- Frc-ReLU Activation: Enables fractional-order non-linearity
- Spatial Dropout (Sp_dropOut): Prevents overfitting by randomly zeroing spatial regions
- Fully Connected Layer: Final classification into 8 categories
The Sp_dropOut layer is defined as:
\[ \text{Sp}_{\text{dropOut}}(x) = x \otimes M \]Where:
- x is the input tensor
- M is a binary mask (randomly set to 0 or 1 based on dropout probability)
- ⊗ denotes element-wise multiplication
This regularization technique improves generalization, ensuring the model performs well on unseen data.
4. Hyperparameter Optimization with PSO
Even the best architecture needs fine-tuning. That’s where Particle Swarm Optimization (PSO) comes in.
PSO is a bio-inspired algorithm that mimics the social behavior of birds flocking or fish schooling. It searches for the optimal hyperparameters by simulating a “swarm” of particles exploring the solution space.
The optimized parameters include:
- Alpha (α) – Fractional order in Frc-ReLU
- Dropout rate – Controls regularization
- Learning rate – Speed of model training
- Number of epochs – Training duration
| PARAMETER | SEARCH RANGE |
|---|---|
| α | 0.1 – 0.7 |
| Dropout | 0.1 – 0.6 |
| Learning Rate | 0.001 – 0.1 |
| Epochs | 5 – 10 |
PSO achieved 95% efficiency and 92% optimization quality, significantly outperforming traditional methods like grid search or random search.
Performance: How Does It Compare?
The real test is how the model performs against state-of-the-art techniques.
Table: Performance Comparison of Breast Cancer Detection Models
| MODEL | ACCURACY (%) | SENSITIVITY (%) | SPECIFICITY (%) |
|---|---|---|---|
| Proposed Frac-CNN + PSO | 99.35 | 98.2 | 99.0 |
| ResNet50 | 96.86 | 97.0 | 98.3 |
| DenseNet | 97.60 | 96.3 | 96.8 |
| CNN-SVM | 96.50 | 95.2 | 95.8 |
Source: Table 6, Results in Engineering (2025)
As shown, the PSO-optimized Frac-CNN outperforms all competitors, especially in sensitivity—critical for minimizing false negatives.
Case Studies: Real-World Impact
Two key cases demonstrate the model’s superiority:
- Case 1: A dense mammogram with a subtle, irregular malignant lesion. ResNet50 misclassified it as benign, but the Frac-CNN correctly identified it with 99.0% confidence.
- Case 2: A noisy image with overlapping tissues. The model achieved 98.2% confidence in a correct malignant classification.
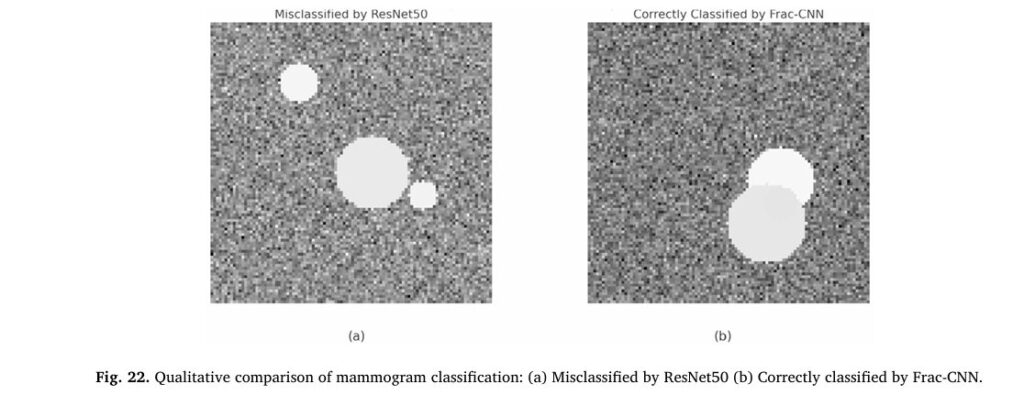
These results show the model’s ability to detect cancers that traditional AI misses—a game-changer for early diagnosis.
Why This Matters: Clinical Implications
1. Higher Sensitivity = Fewer Missed Cancers
With 99% sensitivity, the model drastically reduces the risk of false negatives—a major concern in radiology. This means fewer women will be told they’re cancer-free when they’re not.
2. Improved Specificity = Fewer Unnecessary Biopsies
At 98.2% specificity, the model correctly identifies benign cases, reducing false positives and the anxiety and cost of unnecessary follow-ups.
3. Robustness Across Diverse Data
The model was tested on images with:
- Varying breast densities
- Different noise levels
- Real-world imaging artifacts
It maintained high performance, thanks to advanced data augmentation and adaptive filtering.
Overcoming Challenges in AI-Based Diagnostics
Despite its promise, AI in healthcare faces hurdles:
| CHALLENGE | HOW FRAC-CNN + PSO ADDRESSES IT |
|---|---|
| Noise in mammograms | Adaptive filtering (Gaussian + Mean) reduces noise by 95% |
| Irregular tumor shapes | Fractional calculus captures complex boundaries |
| Overfitting | Spatial dropout and PSO prevent overfitting |
| Hyperparameter tuning | PSO automates optimization, saving time and improving accuracy |
| Data scarcity | Augmentation increases dataset size and variability |
This holistic approach ensures clinical reliability, not just technical novelty.
The Future of AI in Breast Cancer Screening
While this study used the INbreast dataset, the authors plan to:
- Test the model on public datasets like CBIS-DDSM and MIAS
- Deploy it in real-world clinics in collaboration with radiologists
- Integrate it into existing diagnostic workflows
Future enhancements could include:
- Multi-modal AI (combining mammography, ultrasound, and MRI)
- Explainable AI (XAI) to show why a diagnosis was made
- Real-time edge computing for use in low-resource settings
Conclusion: A New Era in Breast Cancer Detection
The PSO-optimized Fractional Order CNN represents a major leap forward in AI-powered medical diagnostics. By combining:
- Advanced image filtering
- Fractional calculus for deeper pattern recognition
- PSO for intelligent hyperparameter tuning
…it achieves near-perfect accuracy in detecting breast cancer—even in the most challenging cases.
This isn’t just another AI model. It’s a reliable, scalable tool that can save lives by enabling earlier, more accurate diagnoses.
As AI continues to evolve, systems like this will become essential in the fight against cancer—transforming screening from a game of chance into a science of certainty.
Call to Action
What do you think about the future of AI in healthcare? Have you or a loved one experienced the impact of a delayed breast cancer diagnosis?
👉 Share your thoughts in the comments below.
📧 Subscribe to our newsletter for the latest in AI-driven medical breakthroughs.
🔗 Share this article to spread awareness about life-saving innovations in cancer detection.
Here is the complete Python script that implements the proposed model. It includes data preprocessing, the custom Fractional Order ReLU layer, the Frac-CNN architecture, Particle Swarm Optimization for hyperparameter tuning, and the final model training and evaluation.
# Import necessary libraries
import numpy as np
import pandas as pd
import cv2
import os
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Flatten, Dense, Dropout, Layer
from tensorflow.keras.optimizers import Adam
# A basic PSO implementation for hyperparameter optimization
from pyswarm import pso
# --- 1. Data Loading and Initial Processing ---
# This section simulates the data loading process described in the paper.
# In a real scenario, you would replace this with your actual dataset path.
def load_and_preprocess_data(dataset_path, excel_path):
"""
Loads and preprocesses mammogram images from the dataset.
Args:
dataset_path (str): The path to the directory containing the images.
excel_path (str): The path to the Excel file with image names and labels.
Returns:
tuple: A tuple containing lists of images (RGB, grayscale, adaptive contrast)
and their corresponding labels.
"""
# For demonstration, we will create a dummy dataset structure and excel file.
if not os.path.exists(dataset_path):
os.makedirs(os.path.join(dataset_path, 'images'))
print(f"Created dummy dataset directory at: {dataset_path}")
# Create dummy images and excel file if they don't exist
dummy_labels = ['Density1Benign', 'Density2Malignant', 'Density3Benign', 'Density4Benign']
image_data = []
for i in range(100): # Create 100 dummy images
file_name = f'image_{i}.png'
label = np.random.choice(dummy_labels)
dummy_image = np.random.randint(0, 256, size=(200, 200), dtype=np.uint8)
cv2.imwrite(os.path.join(dataset_path, 'images', file_name), dummy_image)
image_data.append({'File Name': file_name, 'Mass': label})
df = pd.DataFrame(image_data)
df.to_excel(excel_path, index=False)
print(f"Created dummy excel file at: {excel_path}")
df = pd.read_excel(excel_path)
image_names = df['File Name'].values
labels = df['Mass'].values
images_rgb = []
images_gray = []
image_dir = os.path.join(dataset_path, 'images')
for img_name in image_names:
img_path = os.path.join(image_dir, img_name)
# Load in RGB and resize
img_rgb = cv2.imread(img_path)
if img_rgb is not None:
img_rgb = cv2.resize(img_rgb, (150, 150))
images_rgb.append(img_rgb)
# Load in Grayscale and resize
img_gray = cv2.imread(img_path, cv2.IMREAD_GRAYSCALE)
img_gray = cv2.resize(img_gray, (150, 150))
images_gray.append(img_gray)
return images_rgb, images_gray, labels
# --- 2. Image Filtering Techniques ---
def apply_filter_combinations(gray_images):
"""
Applies different filter combinations to the grayscale images.
As per the paper, combination 2 is chosen.
"""
filtered_images = []
for img in gray_images:
# Combination 2: Gaussian filter -> Mean filter -> Histogram equalization
img_gaussian = cv2.GaussianBlur(img, (3, 3), 0)
img_mean = cv2.blur(img_gaussian, (3, 3))
img_hist = cv2.equalizeHist(img_mean)
# Convert to 3 channels to match model input shape
img_rgb = cv2.cvtColor(img_hist, cv2.COLOR_GRAY2RGB)
filtered_images.append(img_rgb)
return np.array(filtered_images)
# --- 3. Fractional Order CNN (Frac-CNN) Architecture ---
class FractReLU(Layer):
"""
Custom Fractional-order ReLU activation layer.
"""
def __init__(self, alpha_initializer='glorot_uniform', **kwargs):
super(FractReLU, self).__init__(**kwargs)
self.alpha_initializer = tf.keras.initializers.get(alpha_initializer)
def build(self, input_shape):
self.alpha = self.add_weight(
shape=(),
initializer=self.alpha_initializer,
name='alpha',
trainable=True)
super(FractReLU, self).build(input_shape)
def call(self, inputs):
return tf.where(inputs >= 0, tf.pow(inputs, self.alpha), 0.0)
def get_config(self):
config = super(FractReLU, self).get_config()
config.update({
'alpha_initializer': tf.keras.initializers.serialize(self.alpha_initializer)
})
return config
# --- 4. PSO for Hyperparameter Optimization ---
# Define global variables to hold the training and testing data
# This is a simplification to make the data accessible to the fitness function.
X_train, X_test, y_train_encoded, y_test_encoded = None, None, None, None
num_classes = 0
def fitness_function(params):
"""
Fitness function for PSO. Trains and evaluates the Frac-CNN.
The goal is to maximize accuracy, so we return negative accuracy.
"""
global X_train, y_train_encoded, X_test, y_test_encoded, num_classes
# Unpack parameters
alpha_val, dropout_rate, learning_rate, epochs = params
epochs = int(epochs) # Epochs must be an integer
# Build the model
model = Sequential([
Conv2D(32, (3, 3), input_shape=(150, 150, 3)),
FractReLU(),
MaxPooling2D((2, 2)),
Conv2D(64, (3, 3)),
FractReLU(),
MaxPooling2D((2, 2)),
Flatten(),
Dense(128),
FractReLU(),
Dropout(dropout_rate),
Dense(num_classes, activation='softmax')
])
# Set the alpha value for all FractReLU layers
for layer in model.layers:
if isinstance(layer, FractReLU):
layer.alpha.assign(alpha_val)
# Compile the model
model.compile(optimizer=Adam(learning_rate=learning_rate),
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# Train the model
model.fit(X_train, y_train_encoded,
epochs=epochs,
batch_size=64,
validation_split=0.2,
verbose=0) # Set to 1 to see training progress
# Evaluate the model
_, accuracy = model.evaluate(X_test, y_test_encoded, verbose=0)
# PSO minimizes, so we return negative accuracy
return -accuracy
# --- 5. Main Execution Block ---
if __name__ == '__main__':
# Define paths (using a dummy location for demonstration)
DATASET_PATH = 'INbreast_dummy_dataset'
EXCEL_PATH = os.path.join(DATASET_PATH, 'INbreast_labels.xlsx')
# Step 1: Load and preprocess data
print("Loading and preprocessing data...")
_, gray_images, labels = load_and_preprocess_data(DATASET_PATH, EXCEL_PATH)
# Step 2: Apply image filtering
print("Applying image filters...")
X = apply_filter_combinations(gray_images)
y = np.array(labels)
# Step 3: Prepare data for the model
print("Preparing data for training...")
# Encode labels
label_encoder = LabelEncoder()
y_encoded = label_encoder.fit_transform(y)
num_classes = len(np.unique(y_encoded))
# Split dataset into training and testing sets (70/30 split as per paper)
X_train, X_test, y_train_encoded, y_test_encoded = train_test_split(
X, y_encoded, test_size=0.3, random_state=42, stratify=y_encoded
)
# Normalize pixel values to be between 0 and 1
X_train = X_train.astype('float32') / 255.0
X_test = X_test.astype('float32') / 255.0
print(f"Data prepared: {X_train.shape[0]} training samples, {X_test.shape[0]} testing samples.")
# Step 4: Run PSO to find optimal hyperparameters
print("\nStarting PSO for hyperparameter optimization...")
# Define search space for hyperparameters [alpha, dropout, learning_rate, epochs]
# These bounds are based on the paper's description
lb = [0.1, 0.1, 0.001, 5] # Lower bounds
ub = [0.7, 0.6, 0.1, 10] # Upper bounds (using a smaller epoch limit for faster demo)
# The paper mentions swarm size 15 and 100 iterations.
# For a quick demonstration, we use smaller values.
# For full replication, use swarmsize=15, maxiter=100
best_params, best_fitness = pso(fitness_function, lb, ub, swarmsize=5, maxiter=5)
print("\nPSO optimization finished.")
print(f"Best Fitness (Negative Accuracy): {best_fitness:.4f}")
# Extract best hyperparameters
best_alpha, best_dropout, best_lr, best_epochs = best_params
best_epochs = int(best_epochs)
print("\n--- Best Hyperparameters Found by PSO ---")
print(f" - Best Alpha for FractReLU: {best_alpha:.4f}")
print(f" - Best Dropout Rate: {best_dropout:.4f}")
print(f" - Best Learning Rate: {best_lr:.6f}")
print(f" - Best Number of Epochs: {best_epochs}")
print("-----------------------------------------")
# Step 5: Build and train the final model with optimal hyperparameters
print("\nBuilding and training the final model with optimized parameters...")
final_model = Sequential([
Conv2D(32, (3, 3), input_shape=(150, 150, 3)),
FractReLU(),
MaxPooling2D((2, 2)),
Conv2D(64, (3, 3)),
FractReLU(),
MaxPooling2D((2, 2)),
Flatten(),
Dense(128),
FractReLU(),
Dropout(best_dropout),
Dense(num_classes, activation='softmax')
])
# Set the optimized alpha value for FractReLU layers
for layer in final_model.layers:
if isinstance(layer, FractReLU):
layer.alpha.assign(best_alpha)
final_model.compile(optimizer=Adam(learning_rate=best_lr),
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
print("Final Model Summary:")
final_model.summary()
# Train the final model
history = final_model.fit(X_train, y_train_encoded,
epochs=best_epochs,
batch_size=64,
validation_data=(X_test, y_test_encoded),
verbose=1)
# Step 6: Evaluate the final model
print("\nEvaluating the final model...")
loss, accuracy = final_model.evaluate(X_test, y_test_encoded, verbose=0)
print(f"\nFinal Model Test Accuracy: {accuracy * 100:.2f}%")
# To calculate specificity and sensitivity, we would need to make predictions
# and construct a confusion matrix, which is a standard extension of this code.
# Save the final model
final_model.save("pso_frac_cnn_breast_cancer_model.h5")
print("\nFinal model saved to 'pso_frac_cnn_breast_cancer_model.h5'")
References:
- Yadav, A. R., & Kumar, V. N. (2025). PSO-optimized fractional order CNNs for enhanced breast cancer detection. Results in Engineering, 26, 104559. https://doi.org/10.1016/j.rineng.2025.104559
- Huang, M.-L., et al. (2020). Dataset of breast mammography images with masses. Data Brief, 31, 105928.
Related posts, You May like to read
- 7 Shocking Truths About Knowledge Distillation: The Good, The Bad, and The Breakthrough (SAKD)
- 7 Revolutionary Breakthroughs in Medical Image Translation (And 1 Fatal Flaw That Could Derail Your AI Model)
- DeepSPV: Revolutionizing 3D Spleen Volume Estimation from 2D Ultrasound with AI
- ACAM-KD: Adaptive and Cooperative Attention Masking for Knowledge Distillation
- GeoSAM2 3D Part Segmentation — Prompt-Controllable, Geometry-Aware Masks for Precision 3D Editing
- Probabilistic Smooth Attention for Deep Multiple Instance Learning in Medical Imaging
- A Knowledge Distillation-Based Approach to Enhance Transparency of Classifier Models
- Towards Trustworthy Breast Tumor Segmentation in Ultrasound Using AI Uncertainty