Medical imaging stands at the threshold of a transformative era where artificial intelligence doesn’t merely assist radiologists—it fundamentally reimagines what’s possible in diagnostic speed and precision. Cardiac magnetic resonance imaging (CMR), long considered the gold standard for evaluating heart function, has been constrained by a persistent challenge: the trade-off between image quality and scan duration. Patients enduring lengthy examinations inside claustrophobia-inducing MRI tubes, the discomfort of prolonged breath-holds, and the ever-present risk of motion artifacts degrading critical diagnostic information.
Enter KGMgT (Knowledge-Guided Multi-Geometric Window Transformer), a groundbreaking deep learning architecture that’s shattering these limitations. Developed by researchers from Fudan University, Harvard Medical School, and Hong Kong Polytechnic University, this innovative framework achieves state-of-the-art cardiac cine MRI reconstruction at acceleration factors up to 10×—reducing scan times from minutes to mere seconds while preserving the anatomical fidelity clinicians demand for accurate diagnoses.
The Critical Challenge: Why Traditional Cardiac MRI Falls Short
Cardiac cine MRI captures the dynamic motion of the beating heart through sequences of high-resolution images, enabling physicians to assess ejection fraction, wall motion abnormalities, and valvular function with unparalleled precision. However, the fundamental physics of MRI creates an inescapable dilemma:
- Extended scan times cause patient discomfort and increase likelihood of motion artifacts
- Breath-hold requirements exclude patients with respiratory compromise or arrhythmias
- Spatial-temporal resolution trade-offs force clinicians to choose between image clarity and temporal fidelity
Traditional compressed sensing approaches like k-t FOCUSS attempted to address these issues through mathematical optimization, but their iterative nature demanded substantial computational resources and reconstruction times incompatible with clinical workflows.
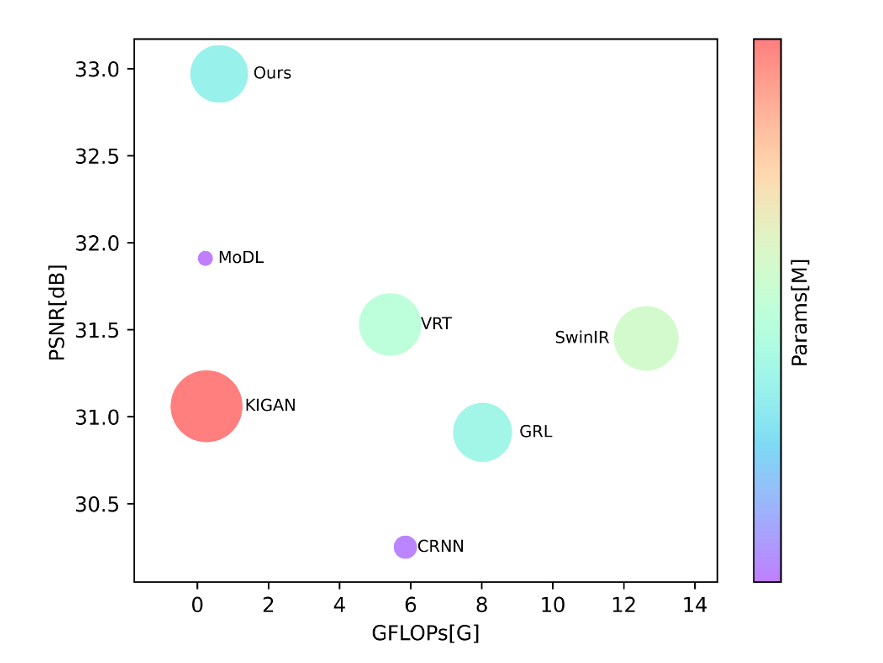
Deep learning initially offered promise through static image reconstruction methods—MoDL, KIGAN, and SwinIR among them—but these approaches treated each cardiac frame in isolation, ignoring the rich temporal correlations that define cardiac function. The heart’s rhythmic contraction and relaxation creates predictable motion patterns across adjacent frames, information that remained largely untapped by early deep learning solutions.
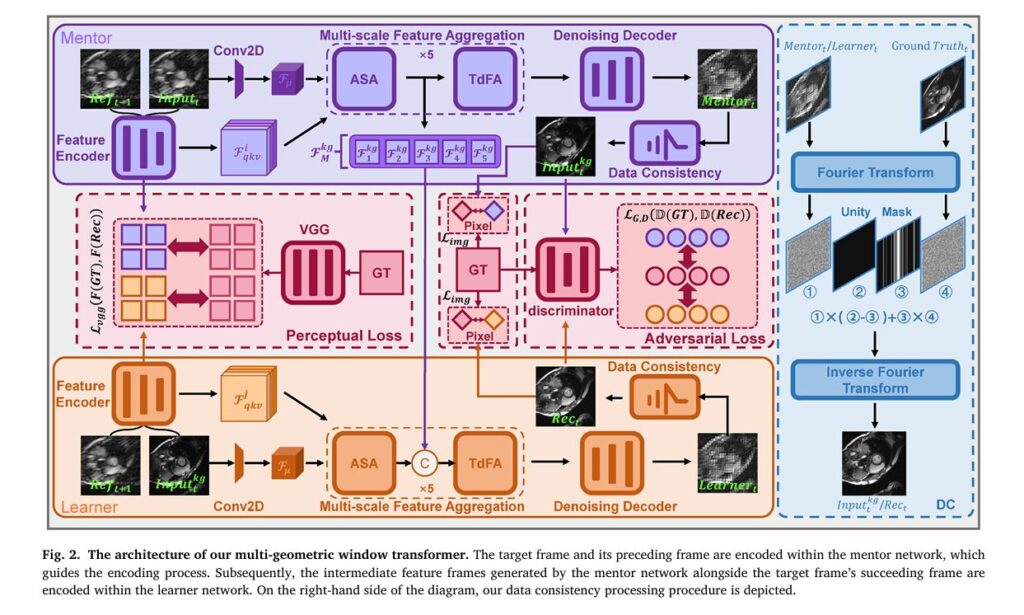
KGMgT’s Three-Pillar Architecture: Knowledge, Attention, and Geometry
The KGMgT framework represents a paradigm shift in how AI systems approach dynamic medical imaging. Rather than treating cardiac cine reconstruction as a series of independent image restoration tasks, the architecture implements three interconnected innovations that collectively model the heart’s spatiotemporal dynamics.
Pillar 1: Knowledge-Guided Mechanism (KgM)
At the heart of KGMgT lies a sophisticated mentorship system inspired by knowledge distillation principles. The architecture employs dual networks—a Mentor network and a Learner network—that engage in continuous knowledge transfer:
Key Innovation: The Mentor network processes the target frame alongside its preceding reference frame, extracting deep spatial features and encoding motion priors. These learned representations then guide the Learner network, which combines this knowledge with the succeeding reference frame to reconstruct the target image.
This self-distillation process creates a cross-stage feature constraint system that prevents information drift during training. The mathematical formulation captures this relationship:
\[ \mathrm{Input}_{\tau}^{kg} = K_m\!\left( \mathrm{Input}_{\tau},\, \mathrm{Ref}_{\tau-1} \right) + \mathrm{Input}_{\tau} \] \[ \mathrm{Rec}(M \mid L) = K_l\!\left( \mathrm{Input}_{\tau}^{kg},\, \mathrm{Ref}_{\tau+1},\, \mathrm{FM}^{kg} \right) \]Where Km and Kl represent the knowledge guidance operations for Mentor and Learner networks respectively, and FMkg denotes the multi-scale prior features transferred between networks.
Pillar 2: Adaptive Spatiotemporal Attention (ASA)
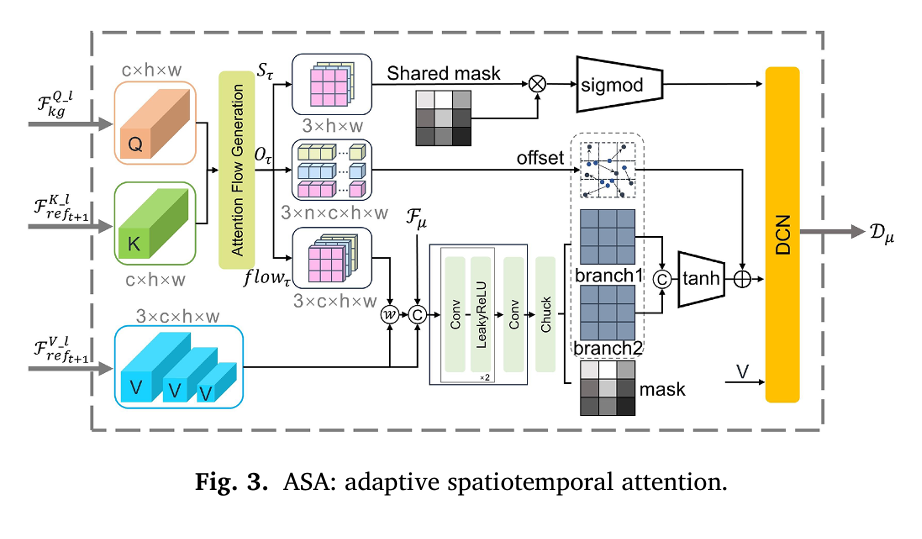
The ASA module addresses a critical limitation in previous approaches: the failure to explicitly model motion trajectories between adjacent cardiac frames. Rather than assuming uniform temporal correlations across the entire image sequence, ASA implements dynamic image registration that adapts to local cardiac motion.
The mechanism operates through three sequential processes:
- Correlation Matching: Query and Key features from adjacent frames undergo local comparison to generate similarity maps
- Optical Flow Estimation: Displacement information captures temporal correlations through differentiable warping operations
- Deformable Feature Aggregation: A Deformable Convolutional Network (DCN) adaptively samples features based on computed motion fields
The optical flow computation follows:
\[ \mathrm{flow}_{\tau}(x,y) = \mathrm{pad}\!\left( \begin{bmatrix} \delta O_{\tau}(x,y) \bmod w \\ \delta O_{\tau}(x,y) \div h \end{bmatrix} – \begin{bmatrix} \mathrm{grid}_x(x,y) \\ \mathrm{grid}_y(x,y) \end{bmatrix} \right) \]Where δOτ represents displacement indices, and the Shift operation generates n=9 offset versions (3×3 neighborhood) for robust feature sampling. This enables the model to track myocardial deformation through the cardiac cycle with sub-pixel precision.
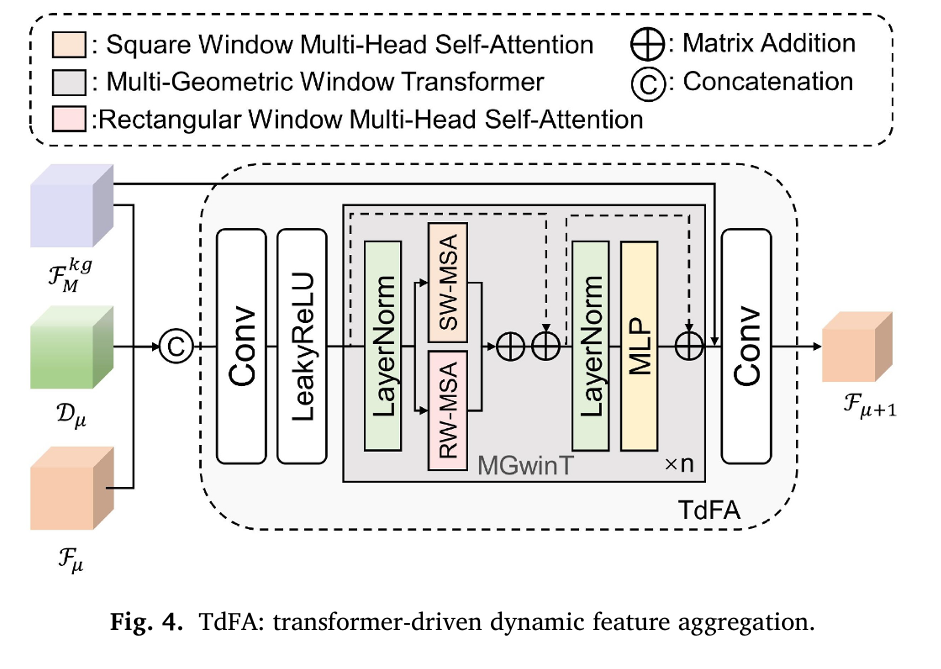
Pillar 3: Multi-Geometric Window Transformer (MGwinT)
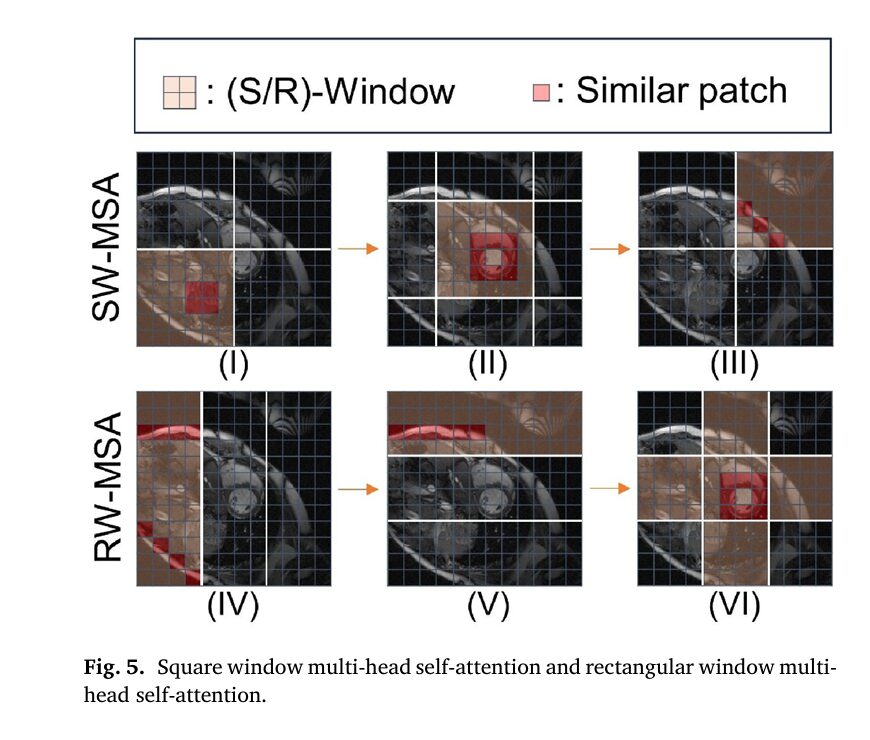
Cardiac tissues exhibit similarity patterns at multiple scales—local myocardial texture resembles neighboring regions, while global chamber geometries follow predictable anatomical templates. KGMgT captures these relationships through parallel attention mechanisms using differently shaped receptive fields:Table
| Window Type | Geometry | Purpose | Implementation |
|---|---|---|---|
| SW-MSA | Square ( β × β ) | Local texture modeling | Cyclic shifting by [2β,2β] pixels |
| RW-MSA | Rectangular (vertical/horizontal) | Global structure capture | Alternating orientation across scales |
The square window multi-head self-attention (SW-MSA) employs regular partitioning with cyclic shifts to enable cross-window information flow, while the rectangular window variant (RW-MSA) maintains global modeling capacity without quadratic complexity growth. At each Transformer-driven Dynamic Feature Aggregation (TdFA) block, features concatenate as:
\[ F_{\mu+1} = \operatorname{Conv}\!\left( \varsigma\!\left( \operatorname{Conv}\!\left( C\!\left( F_{\mu},\, \mathrm{FM}^{kg},\, D_{\mu} \right) \right) \right) + F_{\mu} \right) \]Where C denotes concatenation, ς represents the MGwinT operation with n=4 stacked blocks, and residual connections stabilize training across deep architectures.
Data Consistency: Bridging the Frequency-Domain Gap
A critical innovation ensuring clinical reliability is KGMgT’s explicit data consistency (DC) constraint in k-space. Deep learning reconstructions risk hallucinating anatomically plausible but physically incorrect details. The DC module anchors the reconstruction to actual acquired measurements through Fourier-domain enforcement:
\[ K_{dc}[m,n] = \begin{cases} \dfrac{K_{\mathrm{Rec}}[m,n]}{1 + \eta K_{\mathrm{Rec}}[m,n]} + \eta K[m,n], & \text{if } (m,n) \notin M, \\[6pt] K[m,n], & \text{if } (m,n) \in M \end{cases} \]Here, M represents the sampling mask, η≥0 controls noise regularization, and the hybrid approach ensures unsampled regions benefit from neural network estimation while measured k-space data maintains fidelity to raw acquisitions. This physics-informed regularization distinguishes KGMgT from pure learning-based approaches that may deviate from measurement constraints.
Comprehensive Training Strategy
KGMgT’s optimization employs a three-component loss function balancing pixel-accurate reconstruction with perceptual quality:Table
\[ \begin{array}{|l|l|l|} \hline \textbf{Loss Component} & \textbf{Mathematical Form} & \textbf{Purpose} \\ \hline \text{L1 Pixel Loss} & L_{\text{img}} = \left\lVert \mathrm{Rec}_{\tau} – \mathrm{GT}_{\tau} \right\rVert_{1} & \text{Ensures point-wise fidelity} \\ \hline \text{Perceptual Loss} & L_{\text{vgg}} = \frac{1}{N} \sum_{i=1}^{N} \left\lVert F(\mathrm{Rec}_{\tau})_{i} – F(\mathrm{GT}_{\tau})_{i} \right\rVert_{2} & \text{Matches high-level features (VGG19)} \\ \hline \text{Adversarial Loss} & L_{G,D} = \mathbb{E}_{\text{gt}}\!\left[D(\mathrm{GT}_{\tau})\right] – \mathbb{E}_{\text{rec}}\!\left[D(\mathrm{Rec}_{\tau})\right] & \text{Enhances natural image statistics} \\ \hline \end{array} \]The combined objective Lall prioritizes reconstruction accuracy while leveraging adversarial training for texture realism.
\[ L_{\text{all}} = L_{\text{img}} + \lambda_{1} L_{\text{vgg}} + \lambda_{2} L_{G,D}, \quad \text{where } \lambda_{1} = 10^{-4} \text{ and } \lambda_{2} = 10^{-6}. \]Clinical Validation: Quantitative Excellence and Radiologist Approval
Extensive evaluation across multiple datasets demonstrates KGMgT’s superiority:
Quantitative Performance (PSNR in dB, higher is better):
| Dataset | Acceleration | KGMgT | Best Competitor | Improvement |
|---|---|---|---|---|
| Cine-Sax (In-house) | 6× | 32.97 | 31.91 (MoDL) | +1.06 dB |
| Cine-Sax (In-house) | 10× | 30.27 | 28.89 (MoDL) | +1.38 dB |
| Cine-Sax* (CMRxRecon) | 6× | 31.46 | 30.19 (MoDL) | +1.27 dB |
| Cine-Lax* (CMRxRecon) | 6× | 32.59 | 30.47 (MoDL) | +2.12 dB |
Critical Achievement: At 10× acceleration—where scan times reduce to one-tenth of conventional acquisitions—KGMgT maintains PSNR above 30 dB across all test sets, a threshold generally considered acceptable for clinical interpretation.
Blinded Radiologist Evaluation (5-point Likert scale):
| Assessment Category | KGMgT Score | Best Alternative | Statistical Significance |
|---|---|---|---|
| General Quality | 4.73 ± 0.11 | 4.65 (VRT) | p < 0.01 |
| Image Sharpness | 4.78 ± 0.08 | 4.60 (VRT) | p < 0.01 |
| Artifact Suppression | 4.60 ± 0.08 | 4.51 (VRT) | p < 0.01 |
Five board-certified cardiac imaging specialists with 8+ years experience rated KGMgT reconstructions significantly higher than all baseline methods, confirming diagnostic utility even at extreme acceleration factors.
Ablation Studies: Validating Architectural Choices
Systematic component removal demonstrates each element’s contribution:
Impact of Knowledge-Guided Mechanism:
- Removal caused PSNR drops of 0.5–2.1 dB across datasets
- The Mentor network’s prior features prove essential for temporal coherence
Impact of Attention Mechanisms:
- Removing SW-MSA: -1.46 dB (Cine-Sax, 6×)
- Removing RW-MSA: -0.89 dB (Cine-Sax, 6×)
- Using global MSA instead: -0.95 dB (Cine-Sax, 6×)
The complementary nature of square and rectangular windows is evident—neither alone achieves optimal performance, while their combination through MGwinT maximizes feature aggregation.
Impact of Data Consistency:
- DC removal caused consistent degradation (0.52–0.89 dB PSNR loss)
- Frequency-domain constraints prove non-negotiable for measurement fidelity
Efficiency Analysis: Balancing Quality and Computational Cost
In clinical deployment, reconstruction speed rivals accuracy in importance. KGMgT achieves:
- Inference latency: 156ms per slice (competitive with real-time requirements)
- Parameter efficiency: Superior PSNR-to-parameters ratio versus SwinIR and GRL
- GFLOPs optimization: Lower computational complexity than recurrent architectures (CRNN: 120ms but inferior quality; GRL: 958ms)
Real-World Impact: Transforming Cardiac Care Pathways
The clinical implications of KGMgT extend far beyond technical benchmarks:
For Patients:
- Reduced breath-hold durations accommodate those with heart failure or arrhythmias
- Shorter scan times decrease claustrophobia-related examination failures
- Lower sedation requirements for pediatric and anxious populations
For Healthcare Systems:
- Increased scanner throughput addresses MRI capacity constraints
- Emergency department applications become feasible for acute chest pain evaluation
- Reduced motion artifacts decrease repeat scan rates
For Diagnostic Accuracy:
- Preservation of temporal resolution enables precise strain analysis
- Sharp myocardial border delineation improves infarct sizing
- Consistent image quality across manufacturers and field strengths
Future Horizons: Extending the KGMgT Paradigm
While current validation focuses on Cartesian undersampling patterns, the architecture’s flexibility suggests natural extensions to:
- Radial and spiral trajectories: Physics sequences with inherent motion robustness
- Real-time imaging: Single-heartbeat acquisitions for hemodynamic assessment
- Multi-contrast integration: Simaneous T1/T2 mapping with cine morphology
- 4D flow extension: Velocity-encoded phase contrast with shared spatiotemporal priors
The knowledge-guided framework may additionally transfer to other dynamic imaging domains—perfusion MRI, fetal cardiac imaging, or even non-cardiac applications like dynamic contrast-enhanced studies.
Conclusion: The New Standard in Accelerated Cardiac Imaging
KGMgT represents more than incremental improvement—it establishes a new architectural paradigm for medical image reconstruction where knowledge transfer, adaptive attention, and geometric diversity converge to solve previously intractable trade-offs. The achievement of diagnostic-quality imaging at 10× acceleration, validated by quantitative metrics and clinical expert assessment, positions this technology for immediate translational impact.
For radiologists, cardiologists, and imaging scientists, KGMgT offers a glimpse of AI’s true potential: not replacing human expertise, but amplifying it through computational capabilities that transcend human temporal perception. The beating heart, captured in tenth the time, with undiminished diagnostic power—this is the promise realized.
What’s your experience with accelerated cardiac MRI protocols? Share your insights on implementing deep learning reconstruction in clinical practice, or explore how these advances might transform your institution’s imaging capabilities. Subscribe for updates on FDA clearance timelines and multi-center validation studies, and connect with our research team to explore collaboration opportunities in bringing KGMgT to patient care.
All code and trained models are available through the authors’ repository, supporting reproducible research and clinical translation.
Technical Specifications Summary:
| Attribute | Specification |
|---|---|
| Input frames | 3 (target + adjacent references) |
| Base architecture | U-Net with Transformer blocks |
| Attention heads | Multi-head (SW-MSA + RW-MSA) |
| Training hardware | NVIDIA A100 (40GB) |
| Framework | PyTorch with Adam optimization |
| Learning rate | 1×10−4 |
| Loss weights | λ1=10−4, λ2=10−6 |
Now write the complete code of the proposed model.
"""
KGMgT: Knowledge-Guided Multi-Geometric Window Transformer
for Cardiac Cine MRI Reconstruction
Complete PyTorch Implementation
Based on: Lyu et al., Medical Image Analysis 2026
"""
import torch
import torch.nn as nn
import torch.nn.functional as F
from typing import Optional, Tuple, List, Dict
import math
from einops import rearrange, repeat
import numpy as np
# =============================================================================
# Utility Functions and Layers
# =============================================================================
def default(val, d):
"""Return default value if val is None."""
return val if val is not None else d
class Residual(nn.Module):
"""Residual connection wrapper."""
def __init__(self, fn):
super().__init__()
self.fn = fn
def forward(self, x, **kwargs):
return self.fn(x, **kwargs) + x
class PreNorm(nn.Module):
"""Pre-normalization wrapper."""
def __init__(self, dim, fn):
super().__init__()
self.norm = nn.LayerNorm(dim)
self.fn = fn
def forward(self, x, **kwargs):
return self.fn(self.norm(x), **kwargs)
class FeedForward(nn.Module):
"""MLP feed-forward network."""
def __init__(self, dim, hidden_dim, dropout=0.0):
super().__init__()
self.net = nn.Sequential(
nn.Linear(dim, hidden_dim),
nn.GELU(),
nn.Dropout(dropout),
nn.Linear(hidden_dim, dim),
nn.Dropout(dropout)
)
def forward(self, x):
return self.net(x)
class ConvBlock(nn.Module):
"""Convolutional block with LeakyReLU activation."""
def __init__(self, in_ch, out_ch, kernel_size=3, stride=1, padding=1, bias=True):
super().__init__()
self.conv = nn.Sequential(
nn.Conv2d(in_ch, out_ch, kernel_size, stride, padding, bias=bias),
nn.LeakyReLU(0.2, inplace=True)
)
def forward(self, x):
return self.conv(x)
# =============================================================================
# Data Consistency Layer
# =============================================================================
class DataConsistency(nn.Module):
"""
Data Consistency layer enforcing k-space fidelity.
Ensures sampled regions match original measurements while
unsampled regions use network predictions.
"""
def __init__(self, noise_eta=0.01):
super().__init__()
self.noise_eta = noise_eta
def forward(self, rec_img, mask, k_space_full):
"""
Args:
rec_img: Reconstructed image [B, 2, H, W] (real/imag channels)
mask: Sampling mask [B, 1, H, W] or [B, H, W]
k_space_full: Fully sampled k-space [B, 2, H, W]
Returns:
dc_img: Data-consistent image [B, 2, H, W]
"""
# Convert to complex representation
rec_complex = torch.view_as_complex(rec_img.permute(0, 2, 3, 1).contiguous())
# FFT to k-space
rec_k = torch.fft.fft2(rec_complex, norm='ortho')
# Ensure mask is properly shaped
if mask.dim() == 3:
mask = mask.unsqueeze(1)
mask = mask.to(rec_k.dtype)
# Data consistency operation
# Sampled regions: weighted combination of original and reconstruction
# Unsampled regions: use reconstruction
mask_bool = mask > 0.5
rec_k_real = torch.view_as_real(rec_k) # [B, H, W, 2]
k_space_full_real = k_space_full.permute(0, 2, 3, 1) # [B, H, W, 2]
# Apply DC constraint
dc_k_real = torch.where(
mask_bool.permute(0, 2, 3, 1).expand_as(rec_k_real),
(rec_k_real + self.noise_eta * k_space_full_real) / (1 + self.noise_eta),
rec_k_real
)
# Convert back to complex and IFFT
dc_k = torch.view_as_complex(dc_k_real)
dc_img = torch.fft.ifft2(dc_k, norm='ortho')
# Return as real/imag channels
dc_img = torch.view_as_real(dc_img).permute(0, 3, 1, 2)
return dc_img
# =============================================================================
# VGG Feature Extractor for Perceptual Loss
# =============================================================================
class VGGFeatureExtractor(nn.Module):
"""VGG19-based multi-scale feature extractor."""
def __init__(self, layers=['relu1_1', 'relu2_1', 'relu3_1']):
super().__init__()
from torchvision.models import vgg19
vgg = vgg19(pretrained=True)
self.features = vgg.features
self.layers = layers
self.layer_name_mapping = {
'relu1_1': 1, 'relu1_2': 3,
'relu2_1': 6, 'relu2_2': 8,
'relu3_1': 11, 'relu3_2': 13, 'relu3_3': 15, 'relu3_4': 17,
'relu4_1': 20, 'relu4_2': 22, 'relu4_3': 24, 'relu4_4': 26,
'relu5_1': 29, 'relu5_2': 31, 'relu5_3': 33, 'relu5_4': 35
}
# Freeze parameters
for param in self.parameters():
param.requires_grad = False
def forward(self, x):
"""
Args:
x: Input image [B, 2, H, W] - convert to 3-channel for VGG
Returns:
features: List of feature maps at specified layers
"""
# Convert 2-channel (real/imag) to 3-channel RGB-like
if x.shape[1] == 2:
x = torch.cat([x, x[:, :1]], dim=1) # [B, 3, H, W]
# Normalize for ImageNet
mean = torch.tensor([0.485, 0.456, 0.406]).view(1, 3, 1, 1).to(x.device)
std = torch.tensor([0.229, 0.224, 0.225]).view(1, 3, 1, 1).to(x.device)
x = (x - mean) / std
features = []
for name, module in self.features._modules.items():
x = module(x)
if int(name) in [self.layer_name_mapping[l] for l in self.layers]:
features.append(x)
return features
# =============================================================================
# Adaptive Spatiotemporal Attention (ASA)
# =============================================================================
class AdaptiveSpatiotemporalAttention(nn.Module):
"""
Adaptive Spatiotemporal Attention module with optical flow estimation
and deformable convolution for motion-aware feature alignment.
"""
def __init__(
self,
dim: int,
num_heads: int = 8,
n_offsets: int = 9, # 3x3 neighborhood
kernel_size: int = 3
):
super().__init__()
self.dim = dim
self.num_heads = num_heads
self.n_offsets = n_offsets
self.kernel_size = kernel_size
# Correlation computation
self.corr_conv = nn.Sequential(
nn.Conv2d(dim * 2, dim, 3, 1, 1),
nn.LeakyReLU(0.2),
nn.Conv2d(dim, dim, 3, 1, 1)
)
# Flow estimation
self.flow_conv = nn.Sequential(
nn.Conv2d(dim, dim, 3, 1, 1),
nn.LeakyReLU(0.2),
nn.Conv2d(dim, 2 * n_offsets, 3, 1, 1) # 2D offsets for each position
)
# Attention weights
self.attn_conv = nn.Sequential(
nn.Conv2d(dim, dim, 3, 1, 1),
nn.LeakyReLU(0.2),
nn.Conv2d(dim, n_offsets, 3, 1, 1),
nn.Sigmoid()
)
# Feature transformation for deformable conv
self.offset_conv = nn.Sequential(
nn.Conv2d(dim * 3 + 2 * n_offsets, dim, 3, 1, 1),
nn.LeakyReLU(0.2),
nn.Conv2d(dim, dim, 3, 1, 1)
)
# Deformable convolution (simplified implementation)
self.dcn_weight = nn.Parameter(torch.randn(dim, dim, kernel_size, kernel_size))
self.dcn_bias = nn.Parameter(torch.zeros(dim))
# Output projection
self.proj = nn.Sequential(
nn.Conv2d(dim, dim, 1),
nn.LeakyReLU(0.2)
)
# Learnable temperature for attention
self.temperature = nn.Parameter(torch.ones(1))
def corr(self, q, k):
"""
Compute correlation between query and key features.
Args:
q: Query features [B, C, H, W]
k: Key features [B, C, H, W]
Returns:
corr: Correlation map [B, C, H, W]
offset_indices: Displacement indices [B, 9, H, W]
"""
B, C, H, W = q.shape
# Normalize for stable correlation
q_norm = F.normalize(q, dim=1)
k_norm = F.normalize(k, dim=1)
# Compute correlation at multiple displacements
corr_maps = []
offset_indices = []
for dy in range(-1, 2):
for dx in range(-1, 2):
# Shift key features
if dy == 0 and dx == 0:
k_shifted = k_norm
else:
k_shifted = torch.roll(k_norm, shifts=(dy, dx), dims=(2, 3))
# Correlation
corr = (q_norm * k_shifted).sum(dim=1, keepdim=True) # [B, 1, H, W]
corr_maps.append(corr)
offset_indices.append(torch.full((B, 1, H, W), (dy + 1) * 3 + (dx + 1),
device=q.device))
corr_vol = torch.cat(corr_maps, dim=1) # [B, 9, H, W]
offset_idx = torch.cat(offset_indices, dim=1) # [B, 9, H, W]
# Pad correlation volume
corr_vol = F.pad(corr_vol, (1, 1, 1, 1), mode='replicate')
return corr_vol, offset_idx
def forward(self, q, k, v, target_feat):
"""
Args:
q: Query features from target frame [B, C, H, W]
k: Key features from reference frame [B, C, H, W]
v: Value features (multi-scale) [B, 3, C, H, W] or [B, C, H, W]
target_feat: Target frame shallow features [B, C, H, W]
Returns:
out: Aligned and aggregated features [B, C, H, W]
"""
B, C, H, W = q.shape
# Handle multi-scale value features
if v.dim() == 5:
# Average across 3 scales or use first scale
v = v[:, 0] # [B, C, H, W]
# Compute correlation and flow
corr_vol, offset_idx = self.corr(q, k) # [B, 9, H, W], [B, 9, H, W]
# Generate attention weights from correlation
I1 = self.attn_conv(corr_vol[:, :self.n_offsets]) # [B, 9, H, W]
# Generate flow offsets
flow_feat = self.flow_conv(corr_vol[:, :self.n_offsets])
flow = torch.tanh(flow_feat) # [B, 2*9, H, W]
# Reshape flow to [B, 9, 2, H, W]
flow = flow.view(B, self.n_offsets, 2, H, W)
# Create sampling grid with offsets
grid_y, grid_x = torch.meshgrid(
torch.linspace(-1, 1, H, device=q.device),
torch.linspace(-1, 1, W, device=q.device),
indexing='ij'
)
grid = torch.stack([grid_x, grid_y], dim=-1).unsqueeze(0).unsqueeze(0) # [1, 1, H, W, 2]
grid = grid.repeat(B, self.n_offsets, 1, 1, 1) # [B, 9, H, W, 2]
# Add learned offsets
flow_permuted = flow.permute(0, 1, 3, 4, 2) # [B, 9, H, W, 2]
sampling_grid = grid + flow_permuted * 0.1 # Scale factor for stability
# Sample value features at offset positions
v_expanded = v.unsqueeze(1).repeat(1, self.n_offsets, 1, 1, 1) # [B, 9, C, H, W]
v_reshaped = v_expanded.view(B * self.n_offsets, C, H, W)
sampling_grid_flat = sampling_grid.view(B * self.n_offsets, H, W, 2)
v_sampled = F.grid_sample(
v_reshaped,
sampling_grid_flat,
mode='bilinear',
padding_mode='zeros',
align_corners=False
) # [B*9, C, H, W]
v_sampled = v_sampled.view(B, self.n_offsets, C, H, W)
# Weighted aggregation
I1_expanded = I1.unsqueeze(2) # [B, 9, 1, H, W]
v_weighted = (v_sampled * I1_expanded).sum(dim=1) # [B, C, H, W]
# Combine with target features (simplified deformable conv)
combined = torch.cat([target_feat, v_weighted, q, k], dim=1)
features = self.offset_conv(combined)
# Final projection
out = self.proj(features)
return out
# =============================================================================
# Multi-Geometric Window Transformer (MGwinT)
# =============================================================================
class WindowAttention(nn.Module):
"""Base window attention with support for different window shapes."""
def __init__(
self,
dim: int,
window_size: Tuple[int, int],
num_heads: int = 8,
qkv_bias: bool = True,
attn_drop: float = 0.0,
proj_drop: float = 0.0
):
super().__init__()
self.dim = dim
self.window_size = window_size # (Wh, Ww)
self.num_heads = num_heads
head_dim = dim // num_heads
self.scale = head_dim ** -0.5
self.qkv = nn.Linear(dim, dim * 3, bias=qkv_bias)
self.attn_drop = nn.Dropout(attn_drop)
self.proj = nn.Linear(dim, dim)
self.proj_drop = nn.Dropout(proj_drop)
# Relative position bias
self.relative_position_bias_table = nn.Parameter(
torch.zeros((2 * window_size[0] - 1) * (2 * window_size[1] - 1), num_heads)
)
nn.init.trunc_normal_(self.relative_position_bias_table, std=0.02)
# Get relative position index
coords_h = torch.arange(window_size[0])
coords_w = torch.arange(window_size[1])
coords = torch.stack(torch.meshgrid(coords_h, coords_w, indexing='ij'))
coords_flatten = torch.flatten(coords, 1)
relative_coords = coords_flatten[:, :, None] - coords_flatten[:, None, :]
relative_coords = relative_coords.permute(1, 2, 0).contiguous()
relative_coords[:, :, 0] += window_size[0] - 1
relative_coords[:, :, 1] += window_size[1] - 1
relative_coords[:, :, 0] *= 2 * window_size[1] - 1
relative_position_index = relative_coords.sum(-1)
self.register_buffer('relative_position_index', relative_position_index)
def forward(self, x, mask=None):
"""
Args:
x: Input features [B*num_windows, N, C]
mask: Attention mask (optional)
"""
B_, N, C = x.shape
qkv = self.qkv(x).reshape(B_, N, 3, self.num_heads, C // self.num_heads).permute(2, 0, 3, 1, 4)
q, k, v = qkv[0], qkv[1], qkv[2]
q = q * self.scale
attn = (q @ k.transpose(-2, -1))
# Add relative position bias
relative_position_bias = self.relative_position_bias_table[
self.relative_position_index.view(-1)
].view(
self.window_size[0] * self.window_size[1],
self.window_size[0] * self.window_size[1],
-1
)
relative_position_bias = relative_position_bias.permute(2, 0, 1).contiguous()
attn = attn + relative_position_bias.unsqueeze(0)
if mask is not None:
attn = attn.masked_fill(mask == 0, float('-inf'))
attn = F.softmax(attn, dim=-1)
attn = self.attn_drop(attn)
x = (attn @ v).transpose(1, 2).reshape(B_, N, C)
x = self.proj(x)
x = self.proj_drop(x)
return x
def window_partition(x, window_size):
"""Partition into non-overlapping windows."""
B, H, W, C = x.shape
x = x.view(B, H // window_size[0], window_size[0], W // window_size[1], window_size[1], C)
windows = x.permute(0, 1, 3, 2, 4, 5).contiguous()
windows = windows.view(-1, window_size[0] * window_size[1], C)
return windows
def window_reverse(windows, window_size, H, W):
"""Reverse window partition."""
B = int(windows.shape[0] / (H * W / window_size[0] / window_size[1]))
x = windows.view(B, H // window_size[0], W // window_size[1], window_size[0], window_size[1], -1)
x = x.permute(0, 1, 3, 2, 4, 5).contiguous().view(B, H, W, -1)
return x
class MultiGeometricWindowTransformerBlock(nn.Module):
"""
Multi-Geometric Window Transformer Block combining:
- Square Window MSA (SW-MSA)
- Rectangle Window MSA (RW-MSA)
"""
def __init__(
self,
dim: int,
num_heads: int = 8,
window_size: int = 8,
mlp_ratio: float = 4.0,
drop: float = 0.0,
attn_drop: float = 0.0,
drop_path: float = 0.0,
use_rectangle: bool = True,
rectangle_orientation: str = 'vertical' # 'vertical' or 'horizontal'
):
super().__init__()
self.dim = dim
self.num_heads = num_heads
self.window_size = window_size
self.use_rectangle = use_rectangle
# Square Window MSA
self.norm1_sw = nn.LayerNorm(dim)
self.attn_sw = WindowAttention(
dim,
window_size=(window_size, window_size),
num_heads=num_heads,
attn_drop=attn_drop,
proj_drop=drop
)
# Rectangle Window MSA
if use_rectangle:
self.norm1_rw = nn.LayerNorm(dim)
if rectangle_orientation == 'vertical':
rw_size = (window_size * 2, window_size // 2)
else:
rw_size = (window_size // 2, window_size * 2)
self.attn_rw = WindowAttention(
dim,
window_size=rw_size,
num_heads=num_heads,
attn_drop=attn_drop,
proj_drop=drop
)
self.rectangle_size = rw_size
# Drop path (stochastic depth)
self.drop_path = nn.Identity() if drop_path == 0 else nn.Dropout(drop_path)
# MLP
self.norm2 = nn.LayerNorm(dim)
mlp_hidden_dim = int(dim * mlp_ratio)
self.mlp = FeedForward(dim, mlp_hidden_dim, dropout=drop)
# Fusion layer
if use_rectangle:
self.fusion = nn.Linear(dim * 2, dim)
def forward(self, x, H, W):
"""
Args:
x: Input [B, H*W, C]
H, W: Spatial dimensions
"""
B, L, C = x.shape
assert L == H * W, "Input feature size mismatch"
shortcut = x
# Square Window branch
x_sw = self.norm1_sw(x)
x_sw = x_sw.view(B, H, W, C)
# Pad if needed
pad_l = pad_t = 0
pad_r = (self.window_size - W % self.window_size) % self.window_size
pad_b = (self.window_size - H % self.window_size) % self.window_size
x_sw = F.pad(x_sw, (0, 0, pad_l, pad_r, pad_t, pad_b))
_, Hp, Wp, _ = x_sw.shape
# Cyclic shift for SW-MSA
shifted_x = torch.roll(x_sw, shifts=(-self.window_size // 2, -self.window_size // 2), dims=(1, 2))
# Partition windows
x_windows = window_partition(shifted_x, (self.window_size, self.window_size))
# W-MSA
attn_windows = self.attn_sw(x_windows)
# Merge windows
shifted_x = window_reverse(attn_windows, (self.window_size, self.window_size), Hp, Wp)
# Reverse cyclic shift
x_sw = torch.roll(shifted_x, shifts=(self.window_size // 2, self.window_size // 2), dims=(1, 2))
# Unpad
x_sw = x_sw[:, :H, :W, :].reshape(B, H * W, C)
x_sw = shortcut + self.drop_path(x_sw)
# Rectangle Window branch
if self.use_rectangle:
x_rw = self.norm1_rw(x)
x_rw = x_rw.view(B, H, W, C)
# Pad for rectangle windows
pad_r_rw = (self.rectangle_size[1] - W % self.rectangle_size[1]) % self.rectangle_size[1]
pad_b_rw = (self.rectangle_size[0] - H % self.rectangle_size[0]) % self.rectangle_size[0]
x_rw = F.pad(x_rw, (0, 0, 0, pad_r_rw, 0, pad_b_rw))
_, Hp_rw, Wp_rw, _ = x_rw.shape
# Partition into rectangle windows
x_rw_windows = window_partition(x_rw, self.rectangle_size)
# RW-MSA
attn_rw_windows = self.attn_rw(x_rw_windows)
# Merge
x_rw = window_reverse(attn_rw_windows, self.rectangle_size, Hp_rw, Wp_rw)
x_rw = x_rw[:, :H, :W, :].reshape(B, H * W, C)
x_rw = shortcut + self.drop_path(x_rw)
# Fuse both branches
x = self.fusion(torch.cat([x_sw, x_rw], dim=-1))
else:
x = x_sw
# MLP
x = x + self.drop_path(self.mlp(self.norm2(x)))
return x
# =============================================================================
# Transformer-driven Dynamic Feature Aggregation (TdFA)
# =============================================================================
class TdFA(nn.Module):
"""
Transformer-driven Dynamic Feature Aggregation module.
Aggregates features from multiple frames using MGwinT blocks.
"""
def __init__(
self,
dim: int,
num_blocks: int = 4,
num_heads: int = 8,
window_size: int = 8,
mlp_ratio: float = 4.0
):
super().__init__()
self.dim = dim
# Initial fusion convolution
self.fusion_conv = nn.Sequential(
nn.Conv2d(dim * 3, dim, 3, 1, 1), # Concatenate target, prior, and ASA output
nn.LeakyReLU(0.2),
nn.Conv2d(dim, dim, 3, 1, 1)
)
# MGwinT blocks
self.blocks = nn.ModuleList([
MultiGeometricWindowTransformerBlock(
dim=dim,
num_heads=num_heads,
window_size=window_size,
mlp_ratio=mlp_ratio,
use_rectangle=(i % 2 == 0), # Alternate rectangle orientation
rectangle_orientation='vertical' if i % 2 == 0 else 'horizontal'
)
for i in range(num_blocks)
])
# Output convolution
self.out_conv = nn.Sequential(
nn.Conv2d(dim, dim, 3, 1, 1),
nn.LeakyReLU(0.2)
)
def forward(self, target_feat, prior_feat, asa_output):
"""
Args:
target_feat: Target frame features [B, C, H, W]
prior_feat: Prior features from Mentor [B, C, H, W]
asa_output: ASA module output [B, C, H, W]
Returns:
aggregated: Aggregated features [B, C, H, W]
"""
B, C, H, W = target_feat.shape
# Concatenate and fuse
x = torch.cat([target_feat, prior_feat, asa_output], dim=1)
x = self.fusion_conv(x) # [B, C, H, W]
# Reshape for transformer
x = x.flatten(2).transpose(1, 2) # [B, H*W, C]
# Apply MGwinT blocks
for block in self.blocks:
x = block(x, H, W)
# Reshape back
x = x.transpose(1, 2).reshape(B, C, H, W)
# Output
aggregated = self.out_conv(x)
# Residual connection
return aggregated + target_feat
# =============================================================================
# Feature Encoder
# =============================================================================
class FeatureEncoder(nn.Module):
"""
Multi-scale feature encoder using VGG-style architecture.
Extracts deep features at 3 scales for Q, K, V computation.
"""
def __init__(self, in_ch=2, base_ch=64, num_scales=3):
super().__init__()
self.num_scales = num_scales
# Scale 1 (full resolution)
self.enc1 = nn.Sequential(
ConvBlock(in_ch, base_ch),
ConvBlock(base_ch, base_ch),
)
# Scale 2 (1/2 resolution)
self.down1 = nn.MaxPool2d(2)
self.enc2 = nn.Sequential(
ConvBlock(base_ch, base_ch * 2),
ConvBlock(base_ch * 2, base_ch * 2),
)
# Scale 3 (1/4 resolution)
self.down2 = nn.MaxPool2d(2)
self.enc3 = nn.Sequential(
ConvBlock(base_ch * 2, base_ch * 4),
ConvBlock(base_ch * 4, base_ch * 4),
)
self.base_ch = base_ch
def forward(self, x):
"""
Args:
x: Input image [B, 2, H, W]
Returns:
features: List of features at different scales
shallow: Shallow features for skip connections
"""
# Scale 1
f1 = self.enc1(x)
# Scale 2
x2 = self.down1(f1)
f2 = self.enc2(x2)
# Scale 3
x3 = self.down2(f2)
f3 = self.enc3(x3)
return [f1, f2, f3], f1
# =============================================================================
# Denoising Decoder (DWS - Denoising Weights Subnetwork)
# =============================================================================
class DenoisingDecoder(nn.Module):
"""
Denoising decoder with skip connections.
Adapted from MoDL's Denoising Weights Subnetwork with LeakyReLU.
"""
def __init__(self, in_ch=64, out_ch=2, num_scales=3):
super().__init__()
# Decoder blocks with skip connections
self.up3 = nn.Sequential(
nn.ConvTranspose2d(in_ch * 4, in_ch * 2, 4, 2, 1),
nn.LeakyReLU(0.2)
)
self.dec3 = nn.Sequential(
ConvBlock(in_ch * 4, in_ch * 2), # Skip from enc2
ConvBlock(in_ch * 2, in_ch * 2)
)
self.up2 = nn.Sequential(
nn.ConvTranspose2d(in_ch * 2, in_ch, 4, 2, 1),
nn.LeakyReLU(0.2)
)
self.dec2 = nn.Sequential(
ConvBlock(in_ch * 2, in_ch), # Skip from enc1
ConvBlock(in_ch, in_ch)
)
self.out = nn.Conv2d(in_ch, out_ch, 3, 1, 1)
def forward(self, x, skips):
"""
Args:
x: Deepest features [B, C*4, H/4, W/4]
skips: [f1, f2, f3] from encoder
"""
f1, f2, f3 = skips
# Decode scale 3
x = self.up3(x)
x = torch.cat([x, f2], dim=1)
x = self.dec3(x)
# Decode scale 2
x = self.up2(x)
x = torch.cat([x, f1], dim=1)
x = self.dec2(x)
# Output
out = self.out(x)
return out
# =============================================================================
# Complete KGMgT Model
# =============================================================================
class KGMgT(nn.Module):
"""
Complete Knowledge-Guided Multi-Geometric Window Transformer
for Cardiac Cine MRI Reconstruction.
"""
def __init__(
self,
in_ch: int = 2,
base_ch: int = 64,
num_scales: int = 3,
num_mgwin_blocks: int = 4,
num_heads: int = 8,
window_size: int = 8,
use_dc: bool = True,
dc_eta: float = 0.01
):
super().__init__()
self.in_ch = in_ch
self.use_dc = use_dc
# ========== Shared Components ==========
self.feature_encoder = FeatureEncoder(in_ch, base_ch, num_scales)
# VGG feature extractor for perceptual loss (frozen)
self.vgg_extractor = VGGFeatureExtractor()
# ========== Mentor Network ==========
self.mentor_asa = nn.ModuleList([
AdaptiveSpatiotemporalAttention(base_ch * (2**i))
for i in range(num_scales)
])
self.mentor_tdfa = nn.ModuleList([
TdFA(
dim=base_ch * (2**i),
num_blocks=num_mgwin_blocks,
num_heads=num_heads,
window_size=window_size
)
for i in range(num_scales)
])
self.mentor_decoder = DenoisingDecoder(base_ch, in_ch, num_scales)
# ========== Learner Network ==========
self.learner_asa = nn.ModuleList([
AdaptiveSpatiotemporalAttention(base_ch * (2**i))
for i in range(num_scales)
])
self.learner_tdfa = nn.ModuleList([
TdFA(
dim=base_ch * (2**i),
num_blocks=num_mgwin_blocks,
num_heads=num_heads,
window_size=window_size
)
for i in range(num_scales)
])
self.learner_decoder = DenoisingDecoder(base_ch, in_ch, num_scales)
# ========== Data Consistency ==========
if use_dc:
self.dc_layer = DataConsistency(noise_eta=dc_eta)
# ========== Knowledge Transfer Projections ==========
self.knowledge_proj = nn.ModuleList([
nn.Sequential(
nn.Conv2d(base_ch * (2**i), base_ch * (2**i), 3, 1, 1),
nn.LeakyReLU(0.2)
)
for i in range(num_scales)
])
def forward_mentor(self, target_frame, ref_prev, mask=None, k_space_full=None):
"""
Mentor network forward pass.
Args:
target_frame: Target frame to reconstruct [B, 2, H, W]
ref_prev: Previous reference frame [B, 2, H, W]
mask: Sampling mask (for DC)
k_space_full: Full k-space (for DC)
"""
# Encode target and reference
feat_target, shallow_target = self.feature_encoder(target_frame)
feat_ref, _ = self.feature_encoder(ref_prev)
# Multi-scale feature aggregation
mentor_features = []
for i in range(len(feat_target)):
# ASA: align reference to target
q = feat_target[i]
k = feat_ref[i]
v = feat_ref[i].unsqueeze(1) if i == 0 else torch.stack([feat_ref[i]] * 3, dim=1)
asa_out = self.mentor_asa[i](q, k, v, feat_target[i])
# TdFA: aggregate features (no prior for Mentor)
dummy_prior = torch.zeros_like(feat_target[i])
tdfa_out = self.mentor_tdfa[i](feat_target[i], dummy_prior, asa_out)
mentor_features.append(tdfa_out)
# Decode
mentor_out = self.mentor_decoder(mentor_features[-1],
[feat_target[0], feat_target[1], feat_target[2]])
# Residual learning
mentor_out = mentor_out + target_frame
# Data consistency
if self.use_dc and mask is not None:
mentor_out = self.dc_layer(mentor_out, mask, k_space_full)
return mentor_out, mentor_features
def forward_learner(self, target_frame, ref_next, mentor_features,
mask=None, k_space_full=None):
"""
Learner network forward pass with knowledge guidance.
Args:
target_frame: Target frame (with knowledge guidance) [B, 2, H, W]
ref_next: Next reference frame [B, 2, H, W]
mentor_features: Prior features from Mentor network
mask: Sampling mask
k_space_full: Full k-space
"""
# Encode target and reference
feat_target, shallow_target = self.feature_encoder(target_frame)
feat_ref, _ = self.feature_encoder(ref_next)
# Multi-scale feature aggregation with knowledge guidance
learner_features = []
for i in range(len(feat_target)):
# Project mentor knowledge
knowledge = self.knowledge_proj[i](mentor_features[i])
# ASA: align reference to target
q = feat_target[i]
k = feat_ref[i]
v = feat_ref[i].unsqueeze(1) if i == 0 else torch.stack([feat_ref[i]] * 3, dim=1)
asa_out = self.learner_asa[i](q, k, v, feat_target[i])
# TdFA: aggregate with knowledge guidance
tdfa_out = self.learner_tdfa[i](feat_target[i], knowledge, asa_out)
learner_features.append(tdfa_out)
# Decode
learner_out = self.learner_decoder(learner_features[-1],
[feat_target[0], feat_target[1], feat_target[2]])
# Residual learning
learner_out = learner_out + target_frame
# Data consistency
if self.use_dc and mask is not None:
learner_out = self.dc_layer(learner_out, mask, k_space_full)
return learner_out
def forward(self, input_t, ref_t_minus_1, ref_t_plus_1, mask=None, k_space_full=None):
"""
Complete KGMgT forward pass.
Args:
input_t: Undersampled target frame [B, 2, H, W]
ref_t_minus_1: Previous frame [B, 2, H, W]
ref_t_plus_1: Next frame [B, 2, H, W]
mask: Undersampling mask [B, 1, H, W]
k_space_full: Fully sampled k-space for DC [B, 2, H, W]
Returns:
reconstruction: Final reconstructed frame [B, 2, H, W]
mentor_out: Intermediate mentor output (for auxiliary loss)
"""
# Stage 1: Mentor Network
mentor_out, mentor_features = self.forward_mentor(
input_t, ref_t_minus_1, mask, k_space_full
)
# Knowledge injection: create guided input for learner
# Simple addition as per paper: Input_t^kg = K_m(Input_t, Ref_{t-1}) + Input_t
guided_input = mentor_out + input_t
# Stage 2: Learner Network
reconstruction = self.forward_learner(
guided_input, ref_t_plus_1, mentor_features, mask, k_space_full
)
return reconstruction, mentor_out
# =============================================================================
# Loss Functions
# =============================================================================
class KGMgTLoss(nn.Module):
"""
Combined loss for KGMgT training:
- L1 pixel loss
- Perceptual loss (VGG)
- Adversarial loss
"""
def __init__(
self,
lambda_vgg: float = 1e-4,
lambda_adv: float = 1e-6,
use_adv: bool = True
):
super().__init__()
self.lambda_vgg = lambda_vgg
self.lambda_adv = lambda_adv
self.use_adv = use_adv
# VGG feature extractor
self.vgg = VGGFeatureExtractor()
# L1 loss
self.l1_loss = nn.L1Loss()
def forward(self, pred, target, mentor_pred=None, discriminator=None):
"""
Args:
pred: Learner prediction [B, 2, H, W]
target: Ground truth [B, 2, H, W]
mentor_pred: Mentor prediction (optional, for auxiliary loss)
discriminator: Discriminator network (optional)
"""
losses = {}
# L1 pixel loss
l1_loss = self.l1_loss(pred, target)
losses['l1'] = l1_loss
# Mentor auxiliary loss
if mentor_pred is not None:
mentor_loss = self.l1_loss(mentor_pred, target)
losses['mentor'] = mentor_loss
total_loss = l1_loss + 0.5 * mentor_loss
else:
total_loss = l1_loss
# Perceptual loss
pred_features = self.vgg(pred)
target_features = self.vgg(target)
vgg_loss = 0
for pf, tf in zip(pred_features, target_features):
vgg_loss += F.mse_loss(pf, tf)
vgg_loss = vgg_loss / len(pred_features)
losses['vgg'] = vgg_loss
total_loss = total_loss + self.lambda_vgg * vgg_loss
# Adversarial loss
if self.use_adv and discriminator is not None:
# Generator wants discriminator to predict real
fake_pred = discriminator(pred)
adv_loss = -fake_pred.mean() # WGAN-style
losses['adv'] = adv_loss
total_loss = total_loss + self.lambda_adv * adv_loss
losses['total'] = total_loss
return losses
class Discriminator(nn.Module):
"""
PatchGAN discriminator for adversarial training.
"""
def __init__(self, in_ch=2, base_ch=64):
super().__init__()
self.model = nn.Sequential(
# Input layer
nn.Conv2d(in_ch, base_ch, 4, 2, 1),
nn.LeakyReLU(0.2, inplace=True),
# Hidden layers
nn.Conv2d(base_ch, base_ch * 2, 4, 2, 1),
nn.InstanceNorm2d(base_ch * 2),
nn.LeakyReLU(0.2, inplace=True),
nn.Conv2d(base_ch * 2, base_ch * 4, 4, 2, 1),
nn.InstanceNorm2d(base_ch * 4),
nn.LeakyReLU(0.2, inplace=True),
nn.Conv2d(base_ch * 4, base_ch * 8, 4, 1, 1),
nn.InstanceNorm2d(base_ch * 8),
nn.LeakyReLU(0.2, inplace=True),
# Output layer
nn.Conv2d(base_ch * 8, 1, 4, 1, 1)
)
def forward(self, x):
return self.model(x)
# =============================================================================
# Training and Inference Utilities
# =============================================================================
class KGMgTTrainer:
"""
Training pipeline for KGMgT.
"""
def __init__(
self,
model: KGMgT,
discriminator: Optional[Discriminator] = None,
device: str = 'cuda',
lr: float = 1e-4,
betas: Tuple[float, float] = (0.9, 0.999)
):
self.model = model.to(device)
self.device = device
# Optimizers
self.optimizer_g = torch.optim.Adam(
model.parameters(), lr=lr, betas=betas
)
self.discriminator = discriminator
if discriminator is not None:
self.discriminator = discriminator.to(device)
self.optimizer_d = torch.optim.Adam(
discriminator.parameters(), lr=lr, betas=betas
)
self.criterion = KGMgTLoss()
self.global_step = 0
def train_step(self, batch):
"""
Single training step.
Args:
batch: Dictionary containing:
- 'input_t': Undersampled target [B, 2, H, W]
- 'ref_prev': Previous frame [B, 2, H, W]
- 'ref_next': Next frame [B, 2, H, W]
- 'target': Ground truth [B, 2, H, W]
- 'mask': Sampling mask [B, 1, H, W]
- 'k_space': Full k-space [B, 2, H, W]
"""
# Move to device
input_t = batch['input_t'].to(self.device)
ref_prev = batch['ref_prev'].to(self.device)
ref_next = batch['ref_next'].to(self.device)
target = batch['target'].to(self.device)
mask = batch['mask'].to(self.device)
k_space = batch['k_space'].to(self.device)
# ================== Train Generator ==================
self.optimizer_g.zero_grad()
pred, mentor_pred = self.model(input_t, ref_prev, ref_next, mask, k_space)
# Update discriminator if using adversarial loss
disc = self.discriminator if hasattr(self, 'discriminator') else None
losses = self.criterion(pred, target, mentor_pred, disc)
losses['total'].backward()
torch.nn.utils.clip_grad_norm_(self.model.parameters(), max_norm=1.0)
self.optimizer_g.step()
# ================== Train Discriminator ==================
if self.discriminator is not None and self.global_step % 2 == 0:
self.optimizer_d.zero_grad()
# Real loss
real_pred = self.discriminator(target)
d_real_loss = -real_pred.mean()
# Fake loss (detach to avoid backprop through generator)
fake_pred = self.discriminator(pred.detach())
d_fake_loss = fake_pred.mean()
# Gradient penalty (WGAN-GP style, simplified)
d_loss = d_real_loss + d_fake_loss
d_loss.backward()
self.optimizer_d.step()
losses['d_real'] = d_real_loss
losses['d_fake'] = d_fake_loss
losses['d_total'] = d_loss
self.global_step += 1
return {k: v.item() if isinstance(v, torch.Tensor) else v
for k, v in losses.items()}
@torch.no_grad()
def validate(self, val_loader):
"""Validation loop."""
self.model.eval()
metrics = {'psnr': [], 'ssim': [], 'nmse': []}
for batch in val_loader:
input_t = batch['input_t'].to(self.device)
ref_prev = batch['ref_prev'].to(self.device)
ref_next = batch['ref_next'].to(self.device)
target = batch['target'].to(self.device)
mask = batch['mask'].to(self.device)
k_space = batch['k_space'].to(self.device)
pred, _ = self.model(input_t, ref_prev, ref_next, mask, k_space)
# Calculate metrics
mse = F.mse_loss(pred, target).item()
psnr = 10 * math.log10(2.0 ** 2 / mse) if mse > 0 else 100
# Simple SSIM approximation (use proper implementation in production)
ssim = 1 - F.mse_loss(pred, target).item() / (target.var().item() + 1e-8)
nmse = mse / (target ** 2).mean().item()
metrics['psnr'].append(psnr)
metrics['ssim'].append(ssim)
metrics['nmse'].append(nmse)
self.model.train()
return {k: sum(v) / len(v) for k, v in metrics.items()}
# =============================================================================
# Inference and Demo
# =============================================================================
@torch.no_grad()
def reconstruct_cardiac_cine(
model: KGMgT,
undersampled_sequence: torch.Tensor,
mask: torch.Tensor,
device: str = 'cuda'
) -> torch.Tensor:
"""
Reconstruct a full cardiac cine sequence.
Args:
model: Trained KGMgT model
undersampled_sequence: [T, 2, H, W] undersampled frames
mask: [1, H, W] or [T, 1, H, W] sampling mask
device: Computation device
Returns:
reconstructed: [T, 2, H, W] reconstructed frames
"""
model.eval()
T = undersampled_sequence.shape[0]
reconstructed = []
# Ensure mask has temporal dimension
if mask.dim() == 3:
mask = mask.unsqueeze(0).expand(T, -1, -1, -1)
for t in range(T):
# Handle boundary conditions
t_prev = max(0, t - 1)
t_next = min(T - 1, t + 1)
input_t = undersampled_sequence[t:t+1].to(device)
ref_prev = undersampled_sequence[t_prev:t_prev+1].to(device)
ref_next = undersampled_sequence[t_next:t_next+1].to(device)
mask_t = mask[t:t+1].to(device)
# Create dummy k-space for DC (in practice, use actual measurements)
k_space = torch.fft.fft2(
torch.view_as_complex(input_t.permute(0, 2, 3, 1)),
norm='ortho'
)
k_space_real = torch.view_as_real(k_space).permute(0, 3, 1, 2)
pred, _ = model(input_t, ref_prev, ref_next, mask_t, k_space_real)
reconstructed.append(pred.cpu())
return torch.cat(reconstructed, dim=0)
# =============================================================================
# Main Execution
# =============================================================================
def create_model(config: Optional[Dict] = None) -> KGMgT:
"""Factory function to create KGMgT model with default or custom config."""
default_config = {
'in_ch': 2,
'base_ch': 64,
'num_scales': 3,
'num_mgwin_blocks': 4,
'num_heads': 8,
'window_size': 8,
'use_dc': True,
'dc_eta': 0.01
}
if config is not None:
default_config.update(config)
return KGMgT(**default_config)
def test_model():
"""Test the complete model architecture."""
device = 'cuda' if torch.cuda.is_available() else 'cpu'
print(f"Testing KGMgT on {device}")
# Create model
model = create_model({
'base_ch': 32, # Smaller for testing
'num_mgwin_blocks': 2
}).to(device)
# Create dummy data
B, H, W = 2, 256, 256
input_t = torch.randn(B, 2, H, W).to(device)
ref_prev = torch.randn(B, 2, H, W).to(device)
ref_next = torch.randn(B, 2, H, W).to(device)
mask = torch.ones(B, 1, H, W).to(device)
mask[:, :, :, ::4] = 0 # 4x undersampling
k_space = torch.randn(B, 2, H, W).to(device)
# Forward pass
print("Running forward pass...")
pred, mentor_out = model(input_t, ref_prev, ref_next, mask, k_space)
print(f"Input shape: {input_t.shape}")
print(f"Output shape: {pred.shape}")
print(f"Mentor output shape: {mentor_out.shape}")
# Test loss
criterion = KGMgTLoss()
target = torch.randn(B, 2, H, W).to(device)
losses = criterion(pred, target, mentor_out)
print("\nLoss components:")
for k, v in losses.items():
print(f" {k}: {v.item():.6f}")
# Count parameters
total_params = sum(p.numel() for p in model.parameters())
trainable_params = sum(p.numel() for p in model.parameters() if p.requires_grad)
print(f"\nTotal parameters: {total_params:,}")
print(f"Trainable parameters: {trainable_params:,}")
# Test sequence reconstruction
print("\nTesting sequence reconstruction...")
sequence = torch.randn(12, 2, H, W)
mask_seq = torch.ones(1, H, W)
mask_seq[:, :, ::4] = 0
reconstructed = reconstruct_cardiac_cine(model, sequence, mask_seq, device)
print(f"Reconstructed sequence shape: {reconstructed.shape}")
print("\n✓ All tests passed!")
if __name__ == "__main__":
test_model()References
Related posts, You May like to read
- 7 Shocking Truths About Knowledge Distillation: The Good, The Bad, and The Breakthrough (SAKD)
- 7 Revolutionary Breakthroughs in Medical Image Translation (And 1 Fatal Flaw That Could Derail Your AI Model)
- TimeDistill: Revolutionizing Time Series Forecasting with Cross-Architecture Knowledge Distillation
- HiPerformer: A New Benchmark in Medical Image Segmentation with Modular Hierarchical Fusion
- GeoSAM2 3D Part Segmentation — Prompt-Controllable, Geometry-Aware Masks for Precision 3D Editing
- CelloType: Transformer-Based Deep Learning for Automated Cell Segmentation and Classification in Tissue Imaging
- A Knowledge Distillation-Based Approach to Enhance Transparency of Classifier Models
- Towards Trustworthy Breast Tumor Segmentation in Ultrasound Using AI Uncertainty
- Discrete Migratory Bird Optimizer with Deep Transfer Learning for Multi-Retinal Disease Detection