Introduction: The Growing Challenge of Skin Cancer Diagnosis
Skin cancer remains one of the most prevalent and rapidly increasing forms of cancer worldwide, affecting millions of people annually and placing enormous pressure on healthcare systems. The statistics are sobering: patients diagnosed with melanoma at an early stage enjoy a five-year survival rate of approximately 99%, yet this figure plummets to just 27% when the disease is detected late. This dramatic difference underscores a critical reality in modern medicine—early and accurate diagnosis is quite literally the difference between life and death.
Dermatologists have traditionally relied on visual examination of skin lesions combined with patient history to make diagnostic decisions. They consider not just how a mole or lesion appears, but also crucial contextual information: the patient’s age, whether they smoke, their history of sun exposure, whether the lesion bleeds or itches, and dozens of other clinical indicators. This holistic approach—combining what physicians see with what they know about the patient—has long been the gold standard for diagnosis.
The challenge for artificial intelligence has been replicating this integrated reasoning process. While deep learning systems have achieved remarkable success in analyzing medical images, most approaches treat visual data and patient metadata as separate streams of information, combining them only at the final decision stage. This limitation has prompted researchers to ask a fundamental question: can we teach AI systems to truly fuse these different types of information in the same sophisticated way that experienced clinicians do?
A groundbreaking approach published in the journal Pattern Recognition offers a compelling answer. The Self-contrastive Feature Guidance Based Multidimensional Collaborative Network, or SGMC-Net, represents a paradigm shift in how artificial intelligence processes multimodal medical data, achieving state-of-the-art accuracy in skin disease classification by deeply integrating clinical images with patient metadata throughout the entire feature extraction process.
Understanding the Limitations of Traditional Multimodal Fusion
Before appreciating what makes SGMC-Net revolutionary, it helps to understand how previous systems approached the challenge of combining images with metadata. Traditional multimodal fusion methods typically follow what researchers call “single-level fusion”—they extract features from images using one neural network, process metadata through another pathway, and then concatenate these features just before the final classification layer.
This approach has a fundamental weakness. By keeping the two modalities separate until the very end, these systems miss opportunities to let patient information guide how the network interprets visual features. Consider a concrete example: if a patient’s metadata indicates that a lesion frequently bleeds, this information could help the network pay closer attention to reddish regions in the image. However, with late-stage fusion, this contextual guidance never reaches the image processing pipeline.
The table below illustrates how different fusion strategies compare:
| Fusion Strategy | When Fusion Occurs | Key Limitation |
|---|---|---|
| Simple Concatenation | Final layer only | Ignores modality interactions |
| Attention-based (MetaNet) | Pre-classification | Single-dimensional enhancement |
| MetaBlock | Pre-classification | Channel fusion only |
| SGMC-Net | Throughout extraction | Multidimensional, multi-level |
Key Takeaway: The timing and depth of fusion significantly impact how effectively AI systems can leverage complementary information from different data sources.
The Architecture of SGMC-Net: Building Blocks for Intelligent Fusion
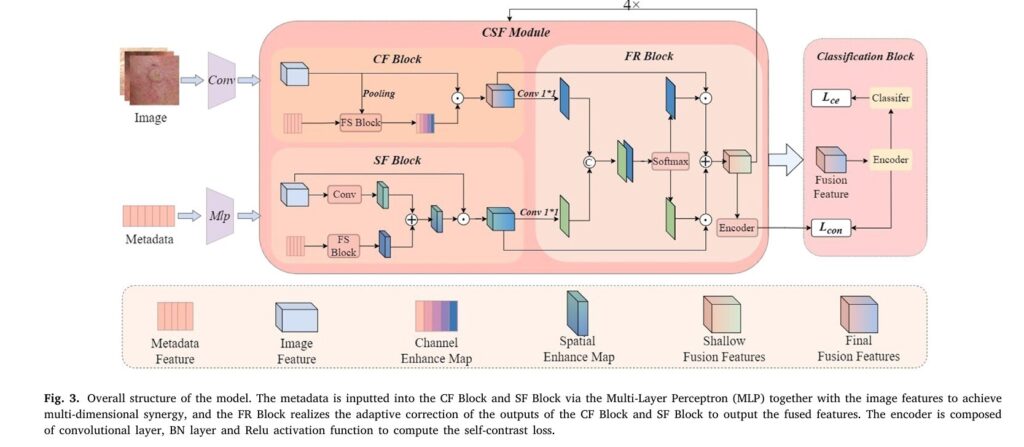
SGMC-Net introduces an elegant solution through what the researchers call “multidimensional collaborative fusion.” Rather than treating metadata as an afterthought, the system weaves patient information into the image analysis process at multiple stages and across multiple dimensions. This is achieved through three specialized components working in concert.
The Channel Fusion Block: Weighting What Matters
In deep learning, image features are organized into channels, with lower-level channels capturing basic patterns like edges and textures, while higher-level channels recognize complex structures like lesion boundaries and color distributions. The Channel Fusion Block (CF Block) allows metadata to dynamically adjust the importance of each channel.
The mathematical operation is elegantly simple. Given image features Ximg ∈ RC×H×W and metadata Xmeta ∈ RC2, the block first maps the metadata to match the number of image channels:
\[ \tilde{X}_{meta} = \sigma(\text{Conv}(X_{meta})) \]where σ\sigma σ represents the sigmoid activation function. The channel-enhanced features are then computed through element-wise multiplication:
\[ \tilde{C}_{img}^i = X_{img}^i \times \tilde{X}_{meta}^i \]This operation allows patient characteristics to selectively amplify or suppress specific feature channels based on clinical relevance.
The Spatial Fusion Block: Pixel-Level Guidance
While channel fusion adjusts which features matter, spatial fusion determines where in the image to focus attention. The Spatial Fusion Block (SF Block) represents a novel contribution, enhancing image features at the individual pixel level based on metadata cues.
The block generates two attention matrices—one derived from the image itself and another from the metadata—then combines them using learnable parameters:
\[ SEM = \sum_{i=1}^{H \times W} (A_{img}^i \times \gamma + A_{meta}^i \times \omega) \]The resulting Spatially Enhanced Map (SEM) guides the network to focus on regions most relevant to the clinical indicators present in the patient’s metadata. If the metadata suggests the lesion has grown over time, for instance, the spatial attention might emphasize boundary regions where growth would be most apparent.
The Feature Rectification Block: Adaptive Balance
Having enhanced features along both channel and spatial dimensions, the network must determine how to optimally combine these complementary representations. The Feature Rectification Block (FR Block) accomplishes this through an adaptive softmax-based weighting mechanism:
\[ R_c^i = \frac{e^{F_c^i}}{e^{F_c^i} + e^{F_s^i}}, \quad R_s^i = \frac{e^{F_s^i}}{e^{F_c^i} + e^{F_s^i}} \]The final fused features emerge from the weighted combination:
\[ F^i = R_c^i \times \tilde{C}_{img}^i + R_s^i \times \tilde{S}_{img}^i \]This adaptive mechanism ensures the network can learn sample-specific fusion strategies, recognizing that different lesions may benefit from different balances of channel versus spatial enhancement.
Self-Contrastive Feature Guidance: A Novel Training Strategy
Beyond its innovative fusion architecture, SGMC-Net introduces a training methodology called self-contrastive feature guidance that addresses a persistent challenge in deep learning: the difficulty of optimizing shallow network layers.
Traditional supervised learning applies loss functions only at the final output layer, requiring error signals to backpropagate through many layers to reach early features. This often results in gradient vanishing, where shallow layers receive weak learning signals and converge slowly. Previous solutions like deep supervision helped by adding auxiliary losses to intermediate layers, but these approaches force shallow features to focus prematurely on classification tasks, potentially interfering with semantic feature extraction.
The self-contrastive approach offers an elegant alternative. Instead of requiring data augmentation like traditional contrastive learning methods, SGMC-Net computes contrastive loss between shallow and deep features within the same batch:
\[ \mathcal{L}_{con}^{self} = \sum_{t=1}^{K-1} \frac{1}{K-1} \cdot \sum_{i \in L} -\log \frac{1}{|P_i|} \sum_{p \in P_i} \frac{\exp(z_i^t \cdot z_p^K / \tau)}{\sum_{a \in A_i} \exp(z_i^t \cdot z_a^K / \tau)} \]In this formulation, features from samples sharing the same disease label are treated as positive pairs, while features from different classes serve as negative pairs. The loss encourages shallow features to align with the semantic structure learned by deeper layers, providing additional supervisory signals without the computational overhead of processing augmented image pairs.
The total training loss combines standard cross-entropy with the self-contrastive term:
\[ \mathcal{L} = \mathcal{L}_{ce} + \delta \cdot \mathcal{L}_{con}^{self} \]where δ is a hyperparameter balancing the two objectives.
Experimental Results: Validating the Approach
The researchers evaluated SGMC-Net on two challenging datasets: PAD-UFES-20, containing clinical images and 21 patient characteristics across six skin lesion categories, and Derm7pt, a seven-point checklist dataset with five diagnostic categories.
The results demonstrated consistent improvements across multiple evaluation metrics:
| Model | Accuracy | Precision | BACC | F1-Score | AUC |
|---|---|---|---|---|---|
| No Metadata | 74.7% | 74.7% | 68.8% | 74.5% | 0.922 |
| Concatenation | 80.4% | 80.0% | 72.4% | 79.9% | 0.938 |
| MetaNet | 78.3% | 77.8% | 71.2% | 77.9% | 0.941 |
| MetaBlock | 73.9% | 72.8% | 64.7% | 72.7% | 0.926 |
| SGMC50 | 83.3% | 83.2% | 75.1% | 83.0% | 0.959 |
The SGMC50 variant improved accuracy by 8.6 percentage points over image-only classification, demonstrating the substantial value of properly integrated multimodal fusion.
Ablation studies further validated each component’s contribution. Removing the Channel Fusion Block decreased accuracy by 7.8%, while eliminating the Spatial Fusion Block caused a 2.4% drop. The self-contrastive feature guidance method proved particularly effective for shallower networks, boosting SGMC18’s accuracy by 5.2 percentage points while adding only 0.6 million parameters.
Clinical Implications and Future Directions
The implications of this research extend beyond academic benchmarks. For clinicians, SGMC-Net demonstrates that AI systems can be designed to reason more like human experts, integrating visual assessment with patient history in a principled, interpretable manner. The attention mechanisms within the spatial and channel fusion blocks could potentially provide insights into which patient characteristics most influence diagnostic decisions for specific lesion types.
Several important considerations warrant attention for clinical deployment:
The performance advantages of multimodal fusion depend heavily on metadata quality and completeness. In real-world settings where patient information may be incomplete or inconsistently recorded, systems must gracefully handle missing data without catastrophic performance degradation. The researchers note that the system removed 358 samples lacking complete metadata information, highlighting this practical consideration.
Additionally, while SGMC-Net achieves impressive aggregate performance, certain disease categories with limited training samples—particularly melanoma with only 43 samples in PAD-UFES-20—remain challenging. Addressing class imbalance through specialized sampling strategies or loss functions represents an important area for continued development.
The Broader Impact on Medical AI
SGMC-Net represents more than an incremental improvement in skin cancer detection; it exemplifies a philosophical shift in how we approach multimodal medical AI. By demonstrating that deep integration of complementary data sources throughout the feature extraction process yields superior results, this research provides a template for other medical imaging applications where clinical context matters.
Consider potential applications in radiology, where imaging findings must be interpreted alongside patient symptoms, laboratory values, and medical history. Or pathology, where tissue samples gain meaning in the context of clinical presentation and genetic markers. The principles underlying SGMC-Net—multidimensional fusion, adaptive feature rectification, and contrastive feature guidance—could enhance AI systems across these domains.
The research also highlights an important insight about the nature of medical diagnosis itself. Experienced physicians do not simply look at images; they synthesize visual information with everything they know about the patient, their risk factors, and the clinical context. AI systems that aspire to match or exceed human diagnostic performance must ultimately learn to perform this same integration, not as an afterthought, but as a fundamental aspect of how they process and understand medical data.
Conclusion: Toward Truly Intelligent Medical AI
The development of SGMC-Net marks a significant milestone in the journey toward AI systems that can genuinely augment clinical decision-making. By introducing multidimensional collaborative fusion and self-contrastive feature guidance, the researchers have demonstrated that thoughtful architectural design can unlock substantially improved performance in medical image analysis.
For healthcare professionals, researchers, and technologists working at the intersection of artificial intelligence and medicine, this work offers both practical advances and conceptual frameworks worth studying closely. The code implementations are becoming increasingly accessible, enabling other researchers to build upon these foundations and extend them to new applications and datasets.
Are you working on medical imaging projects or exploring multimodal AI applications? We encourage you to experiment with these fusion strategies in your own work and share your findings with the community. The path toward AI systems that truly understand medical data in all its complexity requires collaborative effort across disciplines, and advances like SGMC-Net light the way forward.
Featured Image Description: A conceptual diagram showing the SGMC-Net architecture with dermoscopic skin lesion images flowing through multiple fusion stages alongside patient metadata vectors, illustrating how the Channel Fusion Block, Spatial Fusion Block, and Feature Rectification Block work together to produce enhanced diagnostic predictions.
Alt Text: “SGMC-Net multimodal deep learning architecture diagram showing integration of skin lesion images with patient clinical metadata through channel and spatial fusion blocks for improved skin cancer classification accuracy.”
Below is the complete implementation of the Self-contrastive Feature Guidance Based Multidimensional Collaborative Network (SGMC-Net).
"""
SGMC-Net: Self-contrastive Feature Guidance Based Multidimensional Collaborative Network
Paper: Pattern Recognition 156 (2024) 110742
Authors: Feng Li et al.
This implementation provides the complete architecture for skin disease classification
using multimodal fusion of clinical images and patient metadata.
"""
import torch
import torch.nn as nn
import torch.nn.functional as F
from torchvision import models
from typing import Optional, Tuple, List
# =============================================================================
# Feature Selection Block (FS Block) - From authors' previous work MLDF-Net
# Enables dynamic adaptive input of metadata
# =============================================================================
class FSBlock(nn.Module):
"""Feature Selection Block for dynamic metadata processing."""
def __init__(self, meta_dim: int, out_dim: int):
super().__init__()
self.fc = nn.Sequential(
nn.Linear(meta_dim, out_dim),
nn.BatchNorm1d(out_dim),
nn.ReLU(inplace=True)
)
def forward(self, meta: torch.Tensor) -> torch.Tensor:
return self.fc(meta)
# =============================================================================
# Channel Fusion Block (CF Block)
# Generates dynamic responses of image features in channel dimensions based on metadata
# =============================================================================
class CFBlock(nn.Module):
"""
Channel Fusion Block: Enhances image features in the channel dimension using metadata.
The metadata is mapped to match the number of image channels, then used to
weight each channel of the image features via element-wise multiplication.
"""
def __init__(self, img_channels: int, meta_dim: int):
super().__init__()
# Mapping subnet: 1x1 conv equivalent for metadata (implemented as Linear)
# Maps metadata from meta_dim to img_channels
self.meta_mapper = nn.Sequential(
nn.Linear(meta_dim, img_channels),
nn.Sigmoid()
)
def forward(self, x_img: torch.Tensor, x_meta: torch.Tensor) -> torch.Tensor:
"""
Args:
x_img: Image features [B, C, H, W]
x_meta: Metadata features [B, meta_dim]
Returns:
Channel-enhanced image features [B, C, H, W]
"""
B, C, H, W = x_img.shape
# Map metadata to channel weights: [B, meta_dim] -> [B, C]
meta_weights = self.meta_mapper(x_meta) # [B, C]
# Reshape for broadcasting: [B, C] -> [B, C, 1, 1]
meta_weights = meta_weights.view(B, C, 1, 1)
# Channel-wise multiplication: weight each channel by metadata
c_img = x_img * meta_weights
return c_img
# =============================================================================
# Spatial Fusion Block (SF Block)
# Enhances spatial information in images based on metadata
# =============================================================================
class SFBlock(nn.Module):
"""
Spatial Fusion Block: Enhances image features at the pixel level using metadata.
Creates attention matrices from both image and metadata, then combines them
with learnable weights to produce a Spatially Enhanced Map (SEM).
"""
def __init__(self, img_channels: int, meta_dim: int, spatial_size: int = 7):
super().__init__()
# Conv to create image attention matrix: [B, C, H, W] -> [B, 1, H, W]
self.img_conv = nn.Conv2d(img_channels, 1, kernel_size=3, padding=1, stride=1)
# MLP to map metadata to spatial dimensions: [B, meta_dim] -> [B, H*W]
self.meta_mlp = nn.Sequential(
nn.Linear(meta_dim, spatial_size * spatial_size),
nn.ReLU(inplace=True)
)
# Conv to create metadata attention matrix: [B, 1, H, W] -> [B, 1, H, W]
self.meta_conv = nn.Conv2d(1, 1, kernel_size=3, padding=1, stride=1)
# Learnable parameters for weighted combination (gamma and omega)
self.gamma = nn.Parameter(torch.ones(1))
self.omega = nn.Parameter(torch.ones(1))
self.spatial_size = spatial_size
def forward(self, x_img: torch.Tensor, x_meta: torch.Tensor) -> torch.Tensor:
"""
Args:
x_img: Image features [B, C, H, W]
x_meta: Metadata features [B, meta_dim]
Returns:
Spatially-enhanced image features [B, C, H, W]
"""
B, C, H, W = x_img.shape
# Create image attention matrix: [B, C, H, W] -> [B, 1, H, W]
A_img = self.img_conv(x_img)
# Map metadata to spatial dimensions: [B, meta_dim] -> [B, H*W]
meta_spatial = self.meta_mlp(x_meta)
# Reshape and interpolate to match image spatial size: [B, 1, H, W]
meta_spatial = meta_spatial.view(B, 1, self.spatial_size, self.spatial_size)
if H != self.spatial_size or W != self.spatial_size:
meta_spatial = F.interpolate(meta_spatial, size=(H, W), mode='bilinear', align_corners=False)
# Create metadata attention matrix
A_meta = self.meta_conv(meta_spatial)
# Weighted combination to create Spatially Enhanced Map (SEM)
SEM = self.gamma * A_img + self.omega * A_meta
SEM = torch.sigmoid(SEM) # Normalize to [0, 1]
# Apply SEM to input image features
s_img = x_img * SEM
return s_img
# =============================================================================
# Feature Rectification Block (FR Block)
# Adaptive weighted fusion of channel and spatial enhanced features
# =============================================================================
class FRBlock(nn.Module):
"""
Feature Rectification Block: Adaptively fuses channel and spatial enhanced features.
Uses SoftMax-based attention to determine the optimal weighting between
channel-enhanced and spatially-enhanced features.
"""
def __init__(self, channels: int):
super().__init__()
# 1x1 convolutions to generate mapping scores
self.conv_c = nn.Conv2d(channels, 1, kernel_size=1)
self.conv_s = nn.Conv2d(channels, 1, kernel_size=1)
def forward(self, c_img: torch.Tensor, s_img: torch.Tensor) -> torch.Tensor:
"""
Args:
c_img: Channel-enhanced features [B, C, H, W]
s_img: Spatially-enhanced features [B, C, H, W]
Returns:
Fused features [B, C, H, W]
"""
# Generate mapping scores
F_c = self.conv_c(c_img) # [B, 1, H, W]
F_s = self.conv_s(s_img) # [B, 1, H, W]
# Concatenate and apply softmax for adaptive weighting
# Stack along a new dimension for softmax
scores = torch.cat([F_c, F_s], dim=1) # [B, 2, H, W]
weights = F.softmax(scores, dim=1) # [B, 2, H, W]
R_c = weights[:, 0:1, :, :] # [B, 1, H, W]
R_s = weights[:, 1:2, :, :] # [B, 1, H, W]
# Weighted fusion via element-wise addition
fused = R_c * c_img + R_s * s_img
return fused
# =============================================================================
# Channel-Spatial Fusion Module (CSF Module)
# Combines CF Block, SF Block, and FR Block for multi-dimensional synergy
# =============================================================================
class CSFModule(nn.Module):
"""
Channel-Spatial Fusion Module: Achieves multi-level fusion of metadata and image features.
Combines the Channel Fusion Block, Spatial Fusion Block, and Feature Rectification
Block to enable multidimensional collaboration between metadata and images.
"""
def __init__(self, img_channels: int, meta_dim: int, spatial_size: int = 7):
super().__init__()
self.cf_block = CFBlock(img_channels, meta_dim)
self.sf_block = SFBlock(img_channels, meta_dim, spatial_size)
self.fr_block = FRBlock(img_channels)
def forward(self, x_img: torch.Tensor, x_meta: torch.Tensor) -> torch.Tensor:
"""
Args:
x_img: Image features [B, C, H, W]
x_meta: Processed metadata features [B, meta_dim]
Returns:
Fused features [B, C, H, W]
"""
# Channel enhancement
c_img = self.cf_block(x_img, x_meta)
# Spatial enhancement
s_img = self.sf_block(x_img, x_meta)
# Adaptive rectification and fusion
fused = self.fr_block(c_img, s_img)
return fused
# =============================================================================
# Feature Encoder for Self-Contrastive Loss
# Projects features to a lower dimensional space for contrastive learning
# =============================================================================
class ContrastiveEncoder(nn.Module):
"""Encoder for computing self-contrastive loss between shallow and deep features."""
def __init__(self, in_channels: int, out_dim: int = 128):
super().__init__()
self.encoder = nn.Sequential(
nn.Conv2d(in_channels, in_channels, kernel_size=1),
nn.BatchNorm2d(in_channels),
nn.ReLU(inplace=True),
nn.AdaptiveAvgPool2d(1),
nn.Flatten(),
nn.Linear(in_channels, out_dim)
)
def forward(self, x: torch.Tensor) -> torch.Tensor:
return F.normalize(self.encoder(x), dim=1)
# =============================================================================
# SGMC-Net Main Model
# =============================================================================
class SGMCNet(nn.Module):
"""
Self-contrastive Feature Guidance Based Multidimensional Collaborative Network.
This network implements:
1. ResNet backbone for image feature extraction
2. CSF modules for multi-level fusion of metadata and image features
3. Self-contrastive feature guidance for optimizing shallow features
Args:
num_classes: Number of disease categories
meta_dim: Dimension of input metadata
backbone: ResNet variant ('resnet18', 'resnet50', 'resnet152')
pretrained: Whether to use pretrained backbone weights
use_contrastive: Whether to use self-contrastive feature guidance
contrastive_dim: Dimension of contrastive projection space
"""
def __init__(
self,
num_classes: int = 6,
meta_dim: int = 21,
backbone: str = 'resnet18',
pretrained: bool = True,
use_contrastive: bool = True,
contrastive_dim: int = 128
):
super().__init__()
self.use_contrastive = use_contrastive
# Load ResNet backbone
if backbone == 'resnet18':
resnet = models.resnet18(pretrained=pretrained)
channels = [64, 128, 256, 512]
elif backbone == 'resnet50':
resnet = models.resnet50(pretrained=pretrained)
channels = [256, 512, 1024, 2048]
elif backbone == 'resnet152':
resnet = models.resnet152(pretrained=pretrained)
channels = [256, 512, 1024, 2048]
else:
raise ValueError(f"Unsupported backbone: {backbone}")
# Initial convolution layers from ResNet
self.conv1 = resnet.conv1
self.bn1 = resnet.bn1
self.relu = resnet.relu
self.maxpool = resnet.maxpool
# ResNet stages (4 stages for feature extraction)
self.layer1 = resnet.layer1
self.layer2 = resnet.layer2
self.layer3 = resnet.layer3
self.layer4 = resnet.layer4
# Metadata processing: FS Block for dynamic adaptive input
self.fs_block = FSBlock(meta_dim, meta_dim)
# CSF Modules for each ResNet stage (4× as shown in paper Fig. 3)
# Spatial sizes: 56, 28, 14, 7 for 224x224 input
self.csf1 = CSFModule(channels[0], meta_dim, spatial_size=56)
self.csf2 = CSFModule(channels[1], meta_dim, spatial_size=28)
self.csf3 = CSFModule(channels[2], meta_dim, spatial_size=14)
self.csf4 = CSFModule(channels[3], meta_dim, spatial_size=7)
# Contrastive encoders for each stage (for self-contrastive feature guidance)
if use_contrastive:
self.encoder1 = ContrastiveEncoder(channels[0], contrastive_dim)
self.encoder2 = ContrastiveEncoder(channels[1], contrastive_dim)
self.encoder3 = ContrastiveEncoder(channels[2], contrastive_dim)
self.encoder4 = ContrastiveEncoder(channels[3], contrastive_dim)
# Global average pooling and classifier
self.avgpool = nn.AdaptiveAvgPool2d(1)
self.classifier = nn.Linear(channels[3], num_classes)
self.channels = channels
def forward(
self,
img: torch.Tensor,
meta: torch.Tensor,
return_features: bool = False
) -> Tuple[torch.Tensor, Optional[List[torch.Tensor]]]:
"""
Forward pass of SGMC-Net.
Args:
img: Input images [B, 3, 224, 224]
meta: Input metadata [B, meta_dim]
return_features: Whether to return intermediate features for contrastive loss
Returns:
logits: Classification logits [B, num_classes]
features: List of projected features from each stage (if return_features=True)
"""
# Process metadata through FS Block
meta_feat = self.fs_block(meta)
# Initial convolution
x = self.conv1(img)
x = self.bn1(x)
x = self.relu(x)
x = self.maxpool(x)
# Stage 1: ResNet layer + CSF fusion
x = self.layer1(x)
x = self.csf1(x, meta_feat)
f1 = x
# Stage 2: ResNet layer + CSF fusion
x = self.layer2(x)
x = self.csf2(x, meta_feat)
f2 = x
# Stage 3: ResNet layer + CSF fusion
x = self.layer3(x)
x = self.csf3(x, meta_feat)
f3 = x
# Stage 4: ResNet layer + CSF fusion (final fusion features)
x = self.layer4(x)
x = self.csf4(x, meta_feat)
f4 = x
# Global pooling and classification
x = self.avgpool(x)
x = torch.flatten(x, 1)
logits = self.classifier(x)
# Return projected features for contrastive loss if needed
if return_features and self.use_contrastive:
z1 = self.encoder1(f1)
z2 = self.encoder2(f2)
z3 = self.encoder3(f3)
z4 = self.encoder4(f4)
return logits, [z1, z2, z3, z4]
return logits, None
# =============================================================================
# Self-Contrastive Feature Guidance Loss
# =============================================================================
class SelfContrastiveLoss(nn.Module):
"""
Self-Contrastive Feature Guidance Loss.
Computes contrastive loss between shallow and deep features to provide
additional supervisory signals for optimizing shallow layers without
data augmentation.
The loss encourages features from the same class to be similar and
features from different classes to be dissimilar.
"""
def __init__(self, temperature: float = 0.07):
super().__init__()
self.temperature = temperature
def forward(
self,
features: List[torch.Tensor],
labels: torch.Tensor
) -> torch.Tensor:
"""
Compute self-contrastive loss.
Args:
features: List of [z1, z2, z3, z4] projected features from each stage
labels: Ground truth labels [B]
Returns:
Self-contrastive loss scalar
"""
device = features[0].device
batch_size = features[0].shape[0]
K = len(features) # Number of fusion levels
# Get the deepest (final) features as anchor
z_K = features[-1] # [B, dim]
total_loss = 0.0
# Compute contrastive loss for each shallow level against the deep level
for t in range(K - 1):
z_t = features[t] # [B, dim]
# Compute similarity matrix between shallow and deep features
# Each shallow feature is compared against all deep features
sim_matrix = torch.matmul(z_t, z_K.T) / self.temperature # [B, B]
# Create positive mask: samples with same label are positive pairs
labels_equal = labels.unsqueeze(0) == labels.unsqueeze(1) # [B, B]
# For each sample, compute contrastive loss
loss_t = 0.0
for i in range(batch_size):
# Positive samples: same class (excluding self if applicable)
pos_mask = labels_equal[i].clone()
# Number of positive samples
num_pos = pos_mask.sum().item()
if num_pos == 0:
continue
# Compute log-softmax for each row
# Numerator: sum of exp(sim) for positive pairs
# Denominator: sum of exp(sim) for all pairs
exp_sim = torch.exp(sim_matrix[i])
# Sum over positive pairs
pos_sim = (exp_sim * pos_mask.float()).sum()
# Sum over all pairs (negative sample set A_i = all except i)
all_sim = exp_sim.sum()
# Contrastive loss for this sample
loss_i = -torch.log(pos_sim / all_sim + 1e-8)
loss_t += loss_i
loss_t = loss_t / batch_size
total_loss += loss_t / (K - 1)
return total_loss
# =============================================================================
# Combined Loss Function
# =============================================================================
class SGMCLoss(nn.Module):
"""
Combined loss for SGMC-Net training.
Total loss = Cross-Entropy Loss + δ × Self-Contrastive Feature Guidance Loss
"""
def __init__(self, delta: float = 0.1, temperature: float = 0.07):
super().__init__()
self.ce_loss = nn.CrossEntropyLoss()
self.contrastive_loss = SelfContrastiveLoss(temperature)
self.delta = delta
def forward(
self,
logits: torch.Tensor,
labels: torch.Tensor,
features: Optional[List[torch.Tensor]] = None
) -> Tuple[torch.Tensor, torch.Tensor, torch.Tensor]:
"""
Compute combined loss.
Args:
logits: Classification logits [B, num_classes]
labels: Ground truth labels [B]
features: List of projected features (optional, for contrastive loss)
Returns:
total_loss, ce_loss, contrastive_loss
"""
ce = self.ce_loss(logits, labels)
if features is not None:
con = self.contrastive_loss(features, labels)
total = ce + self.delta * con
return total, ce, con
return ce, ce, torch.tensor(0.0)
# =============================================================================
# Model Factory Functions
# =============================================================================
def SGMC18(num_classes: int = 6, meta_dim: int = 21, **kwargs) -> SGMCNet:
"""Create SGMC-Net with ResNet18 backbone."""
return SGMCNet(num_classes, meta_dim, backbone='resnet18', **kwargs)
def SGMC50(num_classes: int = 6, meta_dim: int = 21, **kwargs) -> SGMCNet:
"""Create SGMC-Net with ResNet50 backbone."""
return SGMCNet(num_classes, meta_dim, backbone='resnet50', **kwargs)
def SGMC152(num_classes: int = 6, meta_dim: int = 21, **kwargs) -> SGMCNet:
"""Create SGMC-Net with ResNet152 backbone."""
return SGMCNet(num_classes, meta_dim, backbone='resnet152', **kwargs)
# =============================================================================
# Example Usage and Testing
# =============================================================================
if __name__ == "__main__":
# Test the model
print("=" * 60)
print("Testing SGMC-Net Implementation")
print("=" * 60)
# Configuration for PAD-UFES-20 dataset
batch_size = 4
num_classes = 6 # BCC, SCC, ACK, SEK, MEL, NEV
meta_dim = 21 # 21 patient clinical characteristics
# Create dummy inputs
images = torch.randn(batch_size, 3, 224, 224)
metadata = torch.randn(batch_size, meta_dim)
labels = torch.randint(0, num_classes, (batch_size,))
# Test SGMC18
print("\n[1] Testing SGMC18...")
model18 = SGMC18(num_classes=num_classes, meta_dim=meta_dim, pretrained=False)
logits, features = model18(images, metadata, return_features=True)
print(f" Input image shape: {images.shape}")
print(f" Input metadata shape: {metadata.shape}")
print(f" Output logits shape: {logits.shape}")
print(f" Feature shapes: {[f.shape for f in features]}")
# Count parameters
params = sum(p.numel() for p in model18.parameters()) / 1e6
print(f" Total parameters: {params:.2f}M")
# Test loss computation
criterion = SGMCLoss(delta=0.1)
total_loss, ce_loss, con_loss = criterion(logits, labels, features)
print(f" Total loss: {total_loss.item():.4f}")
print(f" CE loss: {ce_loss.item():.4f}")
print(f" Contrastive loss: {con_loss.item():.4f}")
# Test SGMC50
print("\n[2] Testing SGMC50...")
model50 = SGMC50(num_classes=num_classes, meta_dim=meta_dim, pretrained=False)
logits50, features50 = model50(images, metadata, return_features=True)
print(f" Output logits shape: {logits50.shape}")
params50 = sum(p.numel() for p in model50.parameters()) / 1e6
print(f" Total parameters: {params50:.2f}M")
# Test without contrastive loss
print("\n[3] Testing SGMC18 without contrastive guidance...")
model_no_con = SGMC18(num_classes=num_classes, meta_dim=meta_dim,
pretrained=False, use_contrastive=False)
logits_nc, _ = model_no_con(images, metadata, return_features=False)
print(f" Output logits shape: {logits_nc.shape}")
params_nc = sum(p.numel() for p in model_no_con.parameters()) / 1e6
print(f" Total parameters: {params_nc:.2f}M")
print("\n" + "=" * 60)
print("All tests passed successfully!")
print("=" * 60)References
Related posts, You May like to read
- 7 Shocking Truths About Knowledge Distillation: The Good, The Bad, and The Breakthrough (SAKD)
- 7 Revolutionary Breakthroughs in Medical Image Translation (And 1 Fatal Flaw That Could Derail Your AI Model)
- TimeDistill: Revolutionizing Time Series Forecasting with Cross-Architecture Knowledge Distillation
- HiPerformer: A New Benchmark in Medical Image Segmentation with Modular Hierarchical Fusion
- GeoSAM2 3D Part Segmentation — Prompt-Controllable, Geometry-Aware Masks for Precision 3D Editing
- DGRM: How Advanced AI is Learning to Detect Machine-Generated Text Across Different Domains
- A Knowledge Distillation-Based Approach to Enhance Transparency of Classifier Models
- Towards Trustworthy Breast Tumor Segmentation in Ultrasound Using AI Uncertainty
- Discrete Migratory Bird Optimizer with Deep Transfer Learning for Multi-Retinal Disease Detection