Introduction: The Critical Need for Intelligent Skin Cancer Diagnostics
Skin cancer represents one of the most pervasive and rapidly growing cancer types globally, with incidence rates continuing to climb across all demographics. The primary culprits—DNA damage from ultraviolet (UV) radiation, excessive tanning bed use, and uncontrolled cellular growth—have created a public health imperative for early detection. While melanoma accounts for fewer cases than non-melanoma types like basal cell carcinoma (BCC) and squamous cell carcinoma (SCC), its aggressive nature and potential lethality make accurate identification paramount.
Traditional dermatological diagnosis relies heavily on visual examination and dermoscopy performed by trained specialists. However, this approach carries inherent limitations: subjectivity between practitioners, variability in expertise, and the challenge of detecting subtle morphological differences that distinguish benign from malignant lesions. These limitations become particularly pronounced when dealing with inter-class similarities (where different lesion types look visually alike) and intra-class variations (where the same lesion type presents with different colors, textures, and shapes).
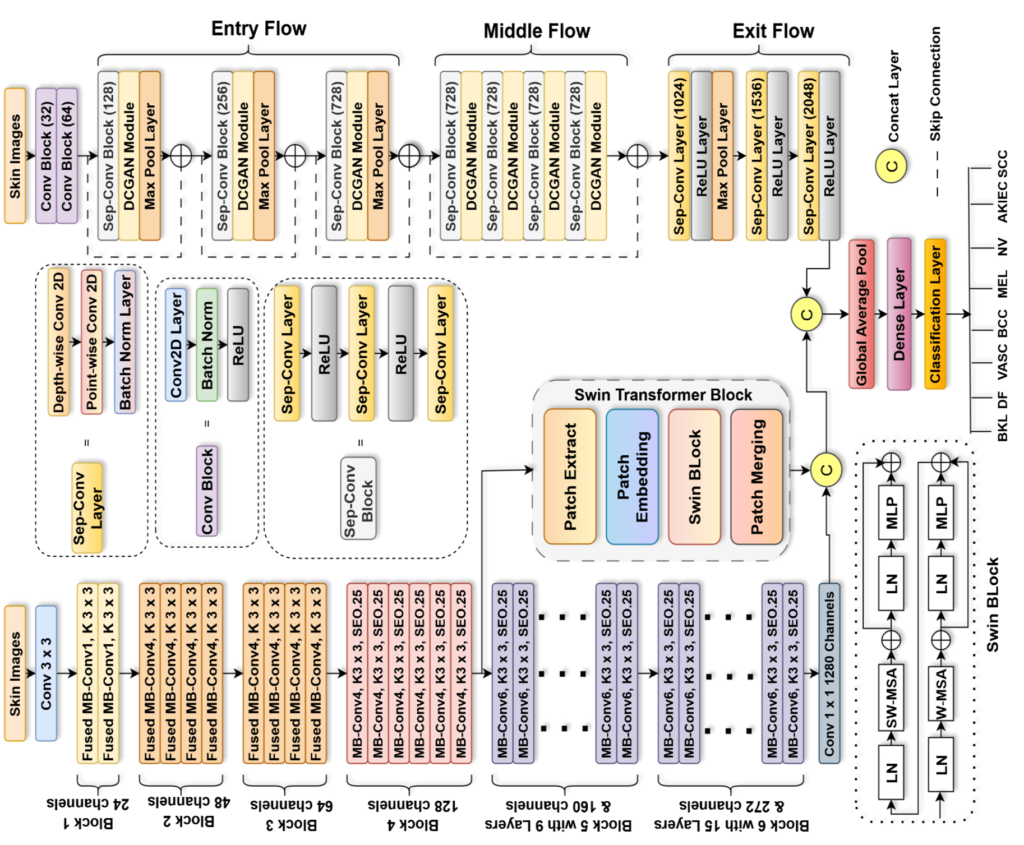
Enter TransXV2S-Net, a groundbreaking hybrid deep learning architecture that represents a paradigm shift in automated skin lesion classification. Developed by researchers at Lahore Leads University, Northeastern University, and Prince Sultan University, this innovative system achieves an remarkable 95.26% accuracy on multi-class skin cancer detection while demonstrating exceptional generalization capabilities across diverse datasets.
Understanding the Architecture: A Three-Pillar Approach to Medical AI
The Hybrid Philosophy: Why Single Models Fall Short
Conventional convolutional neural networks (CNNs), while powerful for local pattern recognition, struggle with long-range dependency modeling and global contextual understanding. Pure transformer architectures like Vision Transformers (ViT) capture global relationships but at prohibitive computational costs and with reduced efficiency for high-resolution medical imagery.
TransXV2S-Net addresses these limitations through a sophisticated multi-branch ensemble architecture that leverages the complementary strengths of three distinct neural network paradigms:
| Component | Primary Function | Key Innovation |
|---|---|---|
| EfficientNetV2S | Feature extraction with compound scaling | Fused-MBConv layers for optimized training |
| Swin Transformer | Global dependency modeling | Hierarchical shifted-window attention |
| Modified Xception + DCGAN | Local contextual refinement | Dual-Contextual Graph Attention Network |
Branch 1: EfficientNetV2S with Embedded Swin Transformer
The first branch harnesses EfficientNetV2S, an evolution of the original EfficientNet architecture that introduces Fused-MBConv layers for the upper network layers. Unlike its predecessor’s 5×5 convolutions, EfficientNetV2S employs 3×3 kernels with optimized expansion ratios, delivering superior training speed without sacrificing representational capacity.
Critically, this branch incorporates a Swin Transformer module at Stage 4, creating a hybrid CNN-transformer pathway. The Swin Transformer processes image patches through a hierarchical structure with shifted window self-attention, enabling efficient computation of global relationships. For a feature map Zl at layer l , the window-based processing follows:
\[ Z_{l}^{\mathrm{win}} \in \mathbb{R}^{\, M_{H_l} \times M_{W_l} \times M^{2} \times D_{l}} \]where M represents the window size and Dl the embedding dimension. The shifted window mechanism alternates between regular and offset window partitions:
\[ Z_{l+1} = \mathrm{SW\text{-}MSA}\!\left( \mathrm{W\text{-}MSA}\!\left( Z_{l} \right) \right) \]This hierarchical attention mechanism, combined with relative positional encoding, allows the model to capture both fine-grained textures and holistic lesion structures—essential for distinguishing morphologically similar conditions.
Branch 2: Modified Xception with DCGAN Integration
The second branch centers on a fundamentally enhanced Xception architecture, where standard separable convolution blocks are augmented with the novel Dual-Contextual Graph Attention Network (DCGAN). This modification represents the paper’s core technical contribution.
The Xception backbone’s depthwise separable convolutions—which decompose standard convolutions into channel-wise spatial operations followed by 1×1 pointwise projections—provide computational efficiency while maintaining feature discriminability. The DCGAN module elevates this capability through three sequential stages:
Stage 1: Context Collector
The Context Collector extracts rich contextual information from input features X∈RH×W×Cin through parallel convolutional pathways:
\[ W_{\psi} = \mathrm{Conv}_{1\times 1}\!\left( X ; K_{\psi} \right) + b_{\psi} \] \[ W_{\phi} = \mathrm{Conv}_{1\times 1}\!\left( X ; K_{\phi} \right) + b_{\phi} \]These intermediate representations undergo dual-path attention processing:
- Channel attention computes cross-spatial importance:
- Spatial attention captures within-channel positional relevance:
The combined context undergoes softmax normalization:
\[ C_{\text{context}}^{\text{mask}} = \operatorname{softmax}\!\left( C_{\text{att}} + S_{\text{att}} \right) \]Stage 2: Graph Convolutional Network with Adaptive Sampling
The GCN stage transforms spatial features into a graph structure where each position becomes a node. To manage computational complexity, adaptive node sampling selects the k most informative nodes based on attention scores:
\[ \boldsymbol{\alpha} = \operatorname{softmax} \!\left( \tanh\!\left( \mathbf{N}\,\mathbf{W}_{\text{att}} \right) \right) \] \[ \mathbf{N}_{\text{sampled}} = \operatorname{TopK} \!\left( \mathbf{N},\, \boldsymbol{\alpha},\, k \right) \]The relationship matrix G between sampled and original nodes enables efficient global context aggregation:
\[ G = N_{\text{sampled}} \cdot N_{T} \in \mathbb{R}^{B \times k \times (H \cdot W)} \]Two-stage graph convolution with residual connections refines features:
\[ H_{1} = \sigma\!\left( N W_{1} + \mathrm{ATN}_{\text{sampled}} \right) \] \[ H_{2} = \sigma\!\left( H_{1} W_{2} + H_{1} \right) \]Stage 3: Context Distributor
The final stage redistributes enhanced contextual information through complementary channel and spatial attention mechanisms, with sigmoid normalization and skip connections preserving original feature information.
Mathematical Framework of Ensemble Fusion
The complete model integrates both branches through learnable fusion weights. For input image x ∈ D ⊂ [0,255]d :
\[ F_{\text{EffSwin}}(x) = \alpha \cdot f_{\text{EffNet}}(x) + \beta \cdot f_{\text{Swin}}(x) \] \[ F_{\text{XceptMod}}(x) = \gamma \cdot f_{\text{Xception}}(x) + \delta \cdot f_{\text{DCGAN}}(x) \] \[ F_{\text{Ensemble}}(x) = \theta \cdot F_{\text{EffSwin}}(x) + \eta \cdot F_{\text{XceptMod}}(x) \]Classification probabilities derive from softmax activation over fully-connected outputs:
\[ y^{k} = \frac{\exp\!\left(F_{\text{Ensemble}}(x)_{k}\right)} {\sum_{j=1}^{K} \exp\!\left(F_{\text{Ensemble}}(x)_{j}\right)} \]Gray World Standard Deviation: Novel Preprocessing for Clinical Robustness
A frequently overlooked aspect of medical AI performance is image preprocessing standardization. TransXV2S-Net introduces the Gray World Standard Deviation (GWSD) algorithm, a sophisticated color normalization technique specifically designed for dermoscopic imagery.
The GWSD algorithm operates through four mathematical steps:
- item Mean intensity calculation per channel:
- item Standard deviation computation:
- item Contrast scaling factor derivation:
where σG represents the target standard deviation.
- item Intensity transformation with clipping:
This preprocessing pipeline, combined with hair removal algorithms and central cropping, ensures that the model learns from clinically relevant features rather than imaging artifacts. Quantitative evaluation demonstrates GWSD’s superiority with PSNR of 24.2 and SSIM of 0.976, outperforming CLAHE, Wiener filtering, and standard Gray World approaches.
Performance Validation: Benchmarking Against State-of-the-Art
ISIC 2019 Dataset Results
On the challenging 8-class ISIC 2019 benchmark (25,331 original samples expanded to 50,662 with preprocessing), TransXV2S-Net achieves:
| Metric | Score | Clinical Significance |
|---|---|---|
| Accuracy | 95.26% | Correct classification in 19 of 20 cases |
| Precision | 94.47% | Low false-positive rate reduces unnecessary biopsies |
| Recall | 94.30% | Critical for malignant lesion detection |
| F1-Score | 94.32% | Balanced performance across all classes |
| AUC-ROC | 99.62% | Excellent discrimination across thresholds |
Bold takeaways from comparative analysis:
- TransXV2S-Net outperforms EFAM-Net by 1.77 percentage points in accuracy while maintaining superior recall (94.30% vs. 90.37%)
- The architecture achieves 3.93 percentage point improvement in recall over the nearest competitor—critical for minimizing missed melanoma diagnoses
- Unlike competing models showing performance imbalances (e.g., DSC-EDLMGWO at 39.49% recall despite 67.35% accuracy), TransXV2S-Net maintains consistent performance above 94% across all metrics
Cross-Dataset Generalization: HAM10000 Validation
True clinical utility requires generalization beyond training distributions. When evaluated on the external HAM10000 dataset (10,015 samples, 7 classes) without retraining, TransXV2S-Net achieves 95% overall accuracy, with exceptional performance on critical classes:
- Nevus (NV): 99% F1-score
- Basal Cell Carcinoma: 97% F1-score
- Melanoma: 96% F1-score
- Benign Keratosis: 97% F1-score
Important limitation identified: The model exhibits complete failure on Vascular Lesions (VASC) in cross-dataset evaluation—0% precision, recall, and F1-score. This stems from extreme class rarity (1.3% of training data) compounded by visual similarity to hemorrhagic melanomas and dataset distribution shift. This finding underscores that high aggregate accuracy can mask critical per-class failures, emphasizing the need for confidence-based flagging systems in clinical deployment.
Ablation Studies: Quantifying Component Contributions
Systematic component removal validates each architectural element’s contribution:
| Configuration | Accuracy | F1-Score | ROC-AUC | Loss |
|---|---|---|---|---|
| Baseline EfficientNetV2S | 89.72% | 86.35% | 98.75% | 0.45 |
| + Swin Transformer | 90.25% | 87.90% | 98.50% | 0.44 |
| + Xception (dual CNN) | 92.97% | 90.42% | 99.25% | 0.34 |
| Full TransXV2S-Net | 95.26% | 94.32% | 99.62% | 0.22 |
The DCGAN module provides the largest single improvement (+2.29% accuracy), demonstrating that graph-based contextual reasoning substantially enhances discrimination of morphologically similar lesions. The progressive reduction in loss values (0.45 → 0.22) indicates superior model fitting without overfitting.
Clinical Implementation Considerations
Computational Efficiency
With 60.69 million parameters and 142.96 images/second throughput, TransXV2S-Net satisfies real-time clinical requirements (>10 images/second) while maintaining diagnostic accuracy. This positions the architecture between lightweight but less accurate models (DSCIMABNet: 1.21M parameters, 75.29% accuracy) and computationally prohibitive alternatives.
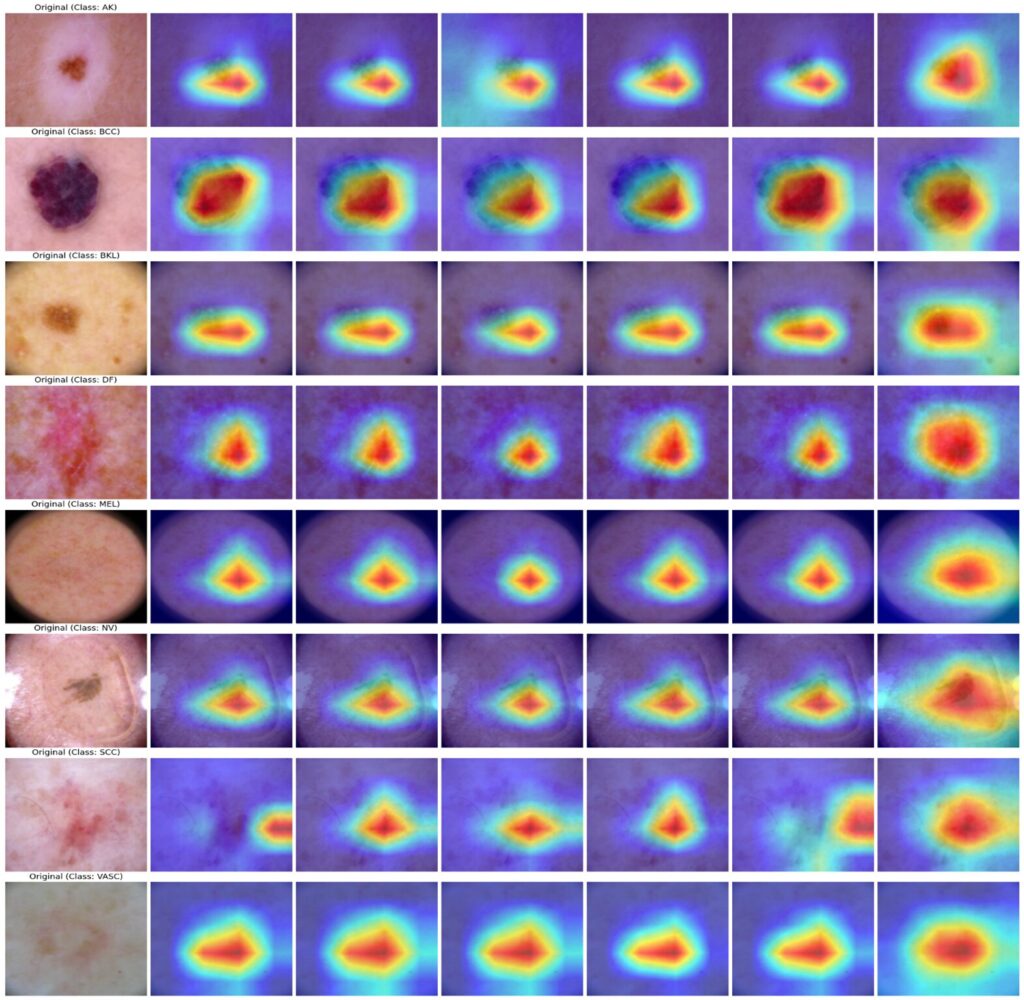
Interpretability Through Attention Visualization
Advanced Grad-CAM techniques (HiResCAM, ScoreCAM, GradCAM++, AblationCAM, EigenCAM, FullGrad) confirm that model decisions align with clinically relevant regions. Heatmap visualizations consistently highlight lesion boundaries, asymmetric structures, and texture irregularities—features dermatologists prioritize during manual examination.
Future Directions and Research Imperatives
The TransXV2S-Net architecture establishes a new foundation for dermatological AI, yet several avenues warrant exploration:
- Minority class enhancement: Implementing class-aware loss functions (focal loss, class-balanced loss) and targeted synthetic data generation through advanced GANs to address extreme imbalance for VASC and similar rare conditions.
- Mobile optimization: Knowledge distillation and neural architecture search to create hardware-efficient variants for point-of-care deployment on smartphones and portable dermoscopes.
- Preprocessing refinement: Extending GWSD to handle challenging illumination conditions and improving hair removal algorithms to preserve diagnostic information while eliminating artifacts.
- Multi-modal integration: Incorporating patient metadata (age, lesion location, family history) and clinical images alongside dermoscopy for comprehensive risk assessment.
Conclusion: Toward Accessible, Reliable Dermatological AI
TransXV2S-Net represents a significant advancement in automated skin lesion classification, demonstrating that thoughtful architectural hybridization—combining CNN efficiency, transformer global modeling, and graph-based relational reasoning—can overcome the persistent challenges of medical image analysis. The 95.26% accuracy on ISIC 2019 and 95% cross-dataset validation on HAM10000, combined with balanced performance across precision and recall metrics, position this system as a viable clinical decision support tool.
However, the complete failure on vascular lesion detection serves as a crucial reminder: AI systems must be deployed with appropriate safeguards, including confidence thresholding and mandatory expert review for low-confidence or rare-class predictions. The goal is not replacement of dermatological expertise but augmentation of diagnostic capacity, particularly in underserved regions with limited specialist access.
As research continues to address minority class performance and computational efficiency, architectures like TransXV2S-Net will play an increasingly vital role in early skin cancer detection, improved patient outcomes, and democratized access to quality dermatological care.
Ready to explore how AI is transforming medical diagnostics?
Subscribe to our research digest for weekly updates on cutting-edge developments in medical AI, or share your thoughts in the comments: What role do you see for automated diagnostic systems in your healthcare experience?
This analysis is based on research published in Knowledge-Based Systems (2026). For the complete technical specifications and implementation details, refer to the original publication: Saeed et al., “TransXV2S-NET: A novel hybrid deep learning architecture with dual-contextual graph attention for multi-class skin lesion classification.”
Below is a complete PyTorch implementation of the TransXV2S-Net model based on the research paper. This is a comprehensive implementation including all components: DCGAN module, modified Xception, EfficientNetV2S with Swin Transformer, and the ensemble architecture.
"""
TransXV2S-Net: Complete PyTorch Implementation
Based on: Saeed et al., Knowledge-Based Systems 2026
"""
import torch
import torch.nn as nn
import torch.nn.functional as F
import math
from typing import Optional, List, Tuple
from timm import create_model
# ============================================================================
# UTILITY FUNCTIONS
# ============================================================================
def drop_path(x, drop_prob: float = 0., training: bool = False):
"""Drop paths (Stochastic Depth) per sample."""
if drop_prob == 0. or not training:
return x
keep_prob = 1 - drop_prob
shape = (x.shape[0],) + (1,) * (x.ndim - 1)
random_tensor = keep_prob + torch.rand(shape, dtype=x.dtype, device=x.device)
random_tensor.floor_()
output = x.div(keep_prob) * random_tensor
return output
class DropPath(nn.Module):
"""Drop paths (Stochastic Depth) per sample."""
def __init__(self, drop_prob: float = 0.):
super().__init__()
self.drop_prob = drop_prob
def forward(self, x):
return drop_path(x, self.drop_prob, self.training)
# ============================================================================
# GRAY WORLD STANDARD DEVIATION (GWSD) PREPROCESSING
# ============================================================================
class GWSDPreprocessor(nn.Module):
"""
Gray World Standard Deviation preprocessing module.
Normalizes color balance and enhances contrast.
"""
def __init__(self, target_std: float = 47.5, target_mean: float = 163.7):
super().__init__()
self.target_std = target_std
self.target_mean = target_mean
def forward(self, x: torch.Tensor) -> torch.Tensor:
"""
Args:
x: Input tensor [B, 3, H, W] in range [0, 1] or [0, 255]
Returns:
Normalized tensor
"""
# Ensure [0, 255] range
if x.max() <= 1.0:
x = x * 255.0
# Process each channel
out = torch.zeros_like(x)
for c in range(3):
channel = x[:, c:c+1, :, :]
mean_c = channel.mean(dim=[2, 3], keepdim=True)
std_c = channel.std(dim=[2, 3], keepdim=True) + 1e-8
# Scaling factor
S_c = self.target_std / std_c
# Transform and clip
transformed = S_c * (channel - mean_c) + self.target_mean
out[:, c:c+1, :, :] = torch.clamp(transformed, 0, 255)
# Normalize back to [0, 1]
return out / 255.0
# ============================================================================
# DUAL-CONTEXTUAL GRAPH ATTENTION NETWORK (DCGAN)
# ============================================================================
class ContextCollector(nn.Module):
"""
Stage 1 of DCGAN: Extracts discriminative local features through
parallel channel and spatial attention.
"""
def __init__(self, in_channels: int, reduction: int = 16):
super().__init__()
self.in_channels = in_channels
# 1x1 convolutions for feature transformation
self.conv_psi = nn.Conv2d(in_channels, in_channels, kernel_size=1)
self.conv_phi = nn.Conv2d(in_channels, in_channels, kernel_size=1)
# Channel attention parameters
self.avg_pool = nn.AdaptiveAvgPool2d(1)
self.max_pool = nn.AdaptiveMaxPool2d(1)
self.mlp = nn.Sequential(
nn.Conv2d(in_channels, in_channels // reduction, 1, bias=False),
nn.ReLU(inplace=True),
nn.Conv2d(in_channels // reduction, in_channels, 1, bias=False)
)
# Spatial attention
self.spatial_conv = nn.Conv2d(2, 1, kernel_size=7, padding=3, bias=False)
def forward(self, x: torch.Tensor) -> torch.Tensor:
B, C, H, W = x.shape
# Transform features
W_psi = self.conv_psi(x) # For channel attention
W_phi = self.conv_phi(x) # For spatial attention
# Channel attention
avg_out = self.mlp(self.avg_pool(W_psi))
max_out = self.mlp(self.max_pool(W_psi))
C_att = torch.sigmoid(avg_out + max_out)
# Spatial attention
avg_spatial = torch.mean(W_phi, dim=1, keepdim=True)
max_spatial, _ = torch.max(W_phi, dim=1, keepdim=True)
spatial_cat = torch.cat([avg_spatial, max_spatial], dim=1)
S_att = torch.sigmoid(self.spatial_conv(spatial_cat))
# Combine attentions
combined = C_att * W_psi + S_att * W_phi
# Softmax normalization across channels
combined_flat = combined.view(B, C, -1)
context_mask = F.softmax(combined_flat, dim=1)
context_mask = context_mask.view(B, C, H, W)
return context_mask * combined
class AdaptiveNodeSampling(nn.Module):
"""
Adaptive node sampling for GCN to reduce computational complexity.
Selects top-k most informative nodes based on attention scores.
"""
def __init__(self, in_channels: int, k: int = 64):
super().__init__()
self.k = k
self.attention_proj = nn.Conv1d(in_channels, 1, kernel_size=1)
def forward(self, x: torch.Tensor) -> Tuple[torch.Tensor, torch.Tensor, torch.Tensor]:
"""
Args:
x: [B, C, N] where N = H*W
Returns:
sampled_nodes: [B, C, k]
attention_scores: [B, N]
indices: [B, k]
"""
B, C, N = x.shape
# Compute attention scores
attn_scores = self.attention_proj(x) # [B, 1, N]
attn_scores = torch.tanh(attn_scores)
attn_scores = F.softmax(attn_scores, dim=-1) # [B, 1, N]
# Top-k sampling
attn_scores_squeeze = attn_scores.squeeze(1) # [B, N]
topk_values, topk_indices = torch.topk(attn_scores_squeeze, self.k, dim=-1)
# Gather sampled nodes
sampled_nodes = torch.stack([
x[b, :, topk_indices[b]] for b in range(B)
], dim=0) # [B, C, k]
return sampled_nodes, attn_scores_squeeze, topk_indices
class GraphConvolutionalNetwork(nn.Module):
"""
Stage 2 of DCGAN: Models global long-range dependencies using GCN
with adaptive node sampling.
"""
def __init__(self, in_channels: int, k: int = 64, num_layers: int = 2):
super().__init__()
self.in_channels = in_channels
self.k = k
self.num_layers = num_layers
# Feature projection
self.proj_conv = nn.Conv2d(in_channels, in_channels, kernel_size=1)
# Adaptive sampling
self.sampler = AdaptiveNodeSampling(in_channels, k)
# GCN layers
self.gcn_weights = nn.ModuleList([
nn.Linear(in_channels, in_channels) for _ in range(num_layers)
])
self.residual_weights = nn.ModuleList([
nn.Linear(in_channels, in_channels) for _ in range(num_layers - 1)
])
self.activation = nn.ReLU(inplace=True)
self.layer_norms = nn.ModuleList([
nn.LayerNorm(in_channels) for _ in range(num_layers)
])
def forward(self, x: torch.Tensor) -> torch.Tensor:
B, C, H, W = x.shape
N = H * W
# Project features
x_proj = self.proj_conv(x) # [B, C, H, W]
# Reshape to graph nodes [B, C, N]
nodes = x_proj.view(B, C, N)
# Adaptive node sampling
sampled_nodes, attn_scores, indices = self.sampler(nodes)
# Compute relationship matrix G: [B, k, N]
G = torch.bmm(sampled_nodes.transpose(1, 2), nodes)
# Scaled dot-product attention
G = G / math.sqrt(C)
A = F.softmax(G, dim=-1) # Attention matrix [B, k, N]
# First GCN layer
H = nodes.transpose(1, 2) # [B, N, C]
sampled_H = sampled_nodes.transpose(1, 2) # [B, k, C]
# Aggregate global context: A^T * sampled_nodes
global_context = torch.bmm(A.transpose(1, 2), sampled_H) # [B, N, C]
H_out = self.gcn_weights[0](H + global_context)
H_out = self.layer_norms[0](H_out)
H_out = self.activation(H_out)
# Second GCN layer with residual
if self.num_layers > 1:
H_residual = H_out
global_context_2 = torch.bmm(A.transpose(1, 2), sampled_H)
H_out = self.gcn_weights[1](H_out + global_context_2) + self.residual_weights[0](H_residual)
H_out = self.layer_norms[1](H_out)
H_out = self.activation(H_out)
# Reshape back to spatial
output = H_out.transpose(1, 2).view(B, C, H, W)
return output
class ContextDistributor(nn.Module):
"""
Stage 3 of DCGAN: Redistributes context-enhanced features.
"""
def __init__(self, in_channels: int, reduction: int = 16):
super().__init__()
# 1x1 convolutions for feature transformation
self.conv_theta = nn.Conv2d(in_channels, in_channels, kernel_size=1)
self.conv_xi = nn.Conv2d(in_channels, in_channels, kernel_size=1)
# Channel attention
self.avg_pool = nn.AdaptiveAvgPool2d(1)
self.max_pool = nn.AdaptiveMaxPool2d(1)
self.mlp = nn.Sequential(
nn.Conv2d(in_channels, in_channels // reduction, 1, bias=False),
nn.ReLU(inplace=True),
nn.Conv2d(in_channels // reduction, in_channels, 1, bias=False)
)
# Spatial attention
self.spatial_conv = nn.Conv2d(2, 1, kernel_size=7, padding=3, bias=False)
def forward(self, x: torch.Tensor, original_input: torch.Tensor) -> torch.Tensor:
# Transform features
W_theta = self.conv_theta(x)
W_xi = self.conv_xi(x)
# Channel attention
avg_out = self.mlp(self.avg_pool(W_theta))
max_out = self.mlp(self.max_pool(W_theta))
C_dist = torch.sigmoid(avg_out + max_out)
# Spatial attention
avg_spatial = torch.mean(W_xi, dim=1, keepdim=True)
max_spatial, _ = torch.max(W_xi, dim=1, keepdim=True)
spatial_cat = torch.cat([avg_spatial, max_spatial], dim=1)
S_dist = torch.sigmoid(self.spatial_conv(spatial_cat))
# Combine and normalize
combined = C_dist * W_theta + S_dist * W_xi
D_mask = torch.sigmoid(combined)
# Skip connection with original input
output = D_mask * x + original_input
return output
class DCGANModule(nn.Module):
"""
Complete Dual-Contextual Graph Attention Network module.
Integrates Context Collector, GCN, and Context Distributor.
"""
def __init__(self, in_channels: int, k: int = 64, reduction: int = 16):
super().__init__()
self.context_collector = ContextCollector(in_channels, reduction)
self.gcn = GraphConvolutionalNetwork(in_channels, k, num_layers=2)
self.context_distributor = ContextDistributor(in_channels, reduction)
def forward(self, x: torch.Tensor) -> torch.Tensor:
original = x
# Stage 1: Context Collection
context = self.context_collector(x)
# Stage 2: Graph Convolution
gcn_out = self.gcn(context)
# Stage 3: Context Distribution
enhanced = self.context_distributor(gcn_out, original)
return enhanced
# ============================================================================
# MODIFIED XCEPTION BACKBONE WITH DCGAN
# ============================================================================
class SeparableConv2d(nn.Module):
"""Depthwise separable convolution."""
def __init__(self, in_channels: int, out_channels: int,
kernel_size: int = 3, stride: int = 1, padding: int = 1,
dilation: int = 1, bias: bool = False):
super().__init__()
self.depthwise = nn.Conv2d(
in_channels, in_channels, kernel_size, stride, padding,
dilation=dilation, groups=in_channels, bias=bias
)
self.pointwise = nn.Conv2d(
in_channels, out_channels, 1, 1, 0, bias=bias
)
def forward(self, x: torch.Tensor) -> torch.Tensor:
x = self.depthwise(x)
x = self.pointwise(x)
return x
class XceptionBlock(nn.Module):
"""Xception block with optional DCGAN and skip connection."""
def __init__(self, in_channels: int, out_channels: int,
num_separable: int = 3, stride: int = 1,
use_dcgab: bool = True, k: int = 64):
super().__init__()
self.use_dcgab = use_dcgab
# Separable convolutions
layers = []
current_channels = in_channels
for i in range(num_separable):
if i == num_separable - 1:
# Last layer may downsample
layers.extend([
SeparableConv2d(current_channels, out_channels,
stride=stride, padding=1),
nn.BatchNorm2d(out_channels),
nn.ReLU(inplace=True)
])
else:
layers.extend([
SeparableConv2d(current_channels, out_channels,
stride=1, padding=1),
nn.BatchNorm2d(out_channels),
nn.ReLU(inplace=True)
])
current_channels = out_channels
self.separable_convs = nn.Sequential(*layers)
# DCGAN module
if use_dcgab:
self.dcgab = DCGANModule(out_channels, k=k)
# Skip connection
if in_channels != out_channels or stride != 1:
self.skip = nn.Sequential(
nn.Conv2d(in_channels, out_channels, 1, stride=stride, bias=False),
nn.BatchNorm2d(out_channels)
)
else:
self.skip = nn.Identity()
self.maxpool = nn.MaxPool2d(3, stride=stride, padding=1) if stride > 1 else nn.Identity()
def forward(self, x: torch.Tensor) -> torch.Tensor:
skip = self.skip(x)
out = self.separable_convs(x)
if self.use_dcgab:
out = self.dcgab(out)
out = out + skip
return out
class ModifiedXception(nn.Module):
"""
Modified Xception architecture with DCGAN integration.
Entry flow, middle flow, and exit flow as described in paper.
"""
def __init__(self, num_classes: int = 8, k: int = 64):
super().__init__()
# ========== Entry Flow ==========
self.entry_flow = nn.Sequential(
# Initial conv layers
nn.Conv2d(3, 32, kernel_size=3, stride=2, padding=1, bias=False),
nn.BatchNorm2d(32),
nn.ReLU(inplace=True),
nn.Conv2d(32, 64, kernel_size=3, stride=1, padding=1, bias=False),
nn.BatchNorm2d(64),
nn.ReLU(inplace=True),
# Entry blocks with DCGAN
XceptionBlock(64, 128, num_separable=3, stride=2, use_dcgab=True, k=k),
XceptionBlock(128, 256, num_separable=3, stride=2, use_dcgab=True, k=k),
XceptionBlock(256, 728, num_separable=3, stride=2, use_dcgab=True, k=k),
)
# ========== Middle Flow ==========
self.middle_flow = nn.Sequential(*[
XceptionBlock(728, 728, num_separable=3, stride=1, use_dcgab=True, k=k)
for _ in range(4)
])
# ========== Exit Flow ==========
self.exit_flow = nn.Sequential(
# Final separable blocks without DCGAN for efficiency
nn.Conv2d(728, 1024, kernel_size=5, stride=1, padding=2, bias=False),
nn.BatchNorm2d(1024),
nn.ReLU(inplace=True),
nn.MaxPool2d(kernel_size=3, stride=2, padding=1),
nn.Conv2d(1024, 1536, kernel_size=5, stride=1, padding=2, bias=False),
nn.BatchNorm2d(1536),
nn.ReLU(inplace=True),
nn.Conv2d(1536, 2048, kernel_size=9, stride=1, padding=4, bias=False),
nn.BatchNorm2d(2048),
nn.ReLU(inplace=True),
)
self.global_pool = nn.AdaptiveAvgPool2d(1)
self.feature_dim = 2048
def forward(self, x: torch.Tensor) -> torch.Tensor:
x = self.entry_flow(x)
x = self.middle_flow(x)
x = self.exit_flow(x)
x = self.global_pool(x)
x = x.flatten(1)
return x
# ============================================================================
# SWIN TRANSFORMER COMPONENTS
# ============================================================================
class WindowAttention(nn.Module):
"""Window based multi-head self attention (W-MSA)."""
def __init__(self, dim: int, window_size: int, num_heads: int,
qkv_bias: bool = True, attn_drop: float = 0., proj_drop: float = 0.):
super().__init__()
self.dim = dim
self.window_size = window_size
self.num_heads = num_heads
head_dim = dim // num_heads
self.scale = head_dim ** -0.5
self.qkv = nn.Linear(dim, dim * 3, bias=qkv_bias)
self.attn_drop = nn.Dropout(attn_drop)
self.proj = nn.Linear(dim, dim)
self.proj_drop = nn.Dropout(proj_drop)
# Relative position bias table
self.relative_position_bias_table = nn.Parameter(
torch.zeros((2 * window_size - 1) ** 2, num_heads)
)
# Get relative position index
coords_h = torch.arange(window_size)
coords_w = torch.arange(window_size)
coords = torch.stack(torch.meshgrid([coords_h, coords_w], indexing='ij'))
coords_flatten = torch.flatten(coords, 1)
relative_coords = coords_flatten[:, :, None] - coords_flatten[:, None, :]
relative_coords = relative_coords.permute(1, 2, 0).contiguous()
relative_coords[:, :, 0] += window_size - 1
relative_coords[:, :, 1] += window_size - 1
relative_coords[:, :, 0] *= 2 * window_size - 1
relative_position_index = relative_coords.sum(-1)
self.register_buffer("relative_position_index", relative_position_index)
nn.init.trunc_normal_(self.relative_position_bias_table, std=.02)
def forward(self, x: torch.Tensor, mask: Optional[torch.Tensor] = None) -> torch.Tensor:
B, N, C = x.shape
qkv = self.qkv(x).reshape(B, N, 3, self.num_heads, C // self.num_heads).permute(2, 0, 3, 1, 4)
q, k, v = qkv[0], qkv[1], qkv[2]
q = q * self.scale
attn = (q @ k.transpose(-2, -1))
# Add relative position bias
relative_position_bias = self.relative_position_bias_table[
self.relative_position_index.view(-1)
].view(self.window_size ** 2, self.window_size ** 2, -1)
relative_position_bias = relative_position_bias.permute(2, 0, 1).contiguous()
attn = attn + relative_position_bias.unsqueeze(0)
if mask is not None:
attn = attn.masked_fill(mask == 0, float('-inf'))
attn = F.softmax(attn, dim=-1)
attn = self.attn_drop(attn)
x = (attn @ v).transpose(1, 2).reshape(B, N, C)
x = self.proj(x)
x = self.proj_drop(x)
return x
class SwinTransformerBlock(nn.Module):
"""Swin Transformer Block with W-MSA and SW-MSA."""
def __init__(self, dim: int, num_heads: int, window_size: int = 4,
shift_size: int = 0, mlp_ratio: float = 4., drop: float = 0.,
attn_drop: float = 0., drop_path: float = 0.):
super().__init__()
self.dim = dim
self.num_heads = num_heads
self.window_size = window_size
self.shift_size = shift_size
self.norm1 = nn.LayerNorm(dim)
self.attn = WindowAttention(dim, window_size, num_heads,
attn_drop=attn_drop, proj_drop=drop)
self.drop_path = DropPath(drop_path) if drop_path > 0. else nn.Identity()
self.norm2 = nn.LayerNorm(dim)
mlp_hidden_dim = int(dim * mlp_ratio)
self.mlp = nn.Sequential(
nn.Linear(dim, mlp_hidden_dim),
nn.GELU(),
nn.Dropout(drop),
nn.Linear(mlp_hidden_dim, dim),
nn.Dropout(drop)
)
def forward(self, x: torch.Tensor, H: int, W: int) -> torch.Tensor:
B, N, C = x.shape
shortcut = x
x = self.norm1(x)
x = x.view(B, H, W, C)
# Cyclic shift
if self.shift_size > 0:
shifted_x = torch.roll(x, shifts=(-self.shift_size, -self.shift_size), dims=(1, 2))
else:
shifted_x = x
# Partition windows
x_windows = self.window_partition(shifted_x, self.window_size)
x_windows = x_windows.view(-1, self.window_size * self.window_size, C)
# W-MSA/SW-MSA
attn_windows = self.attn(x_windows)
# Merge windows
attn_windows = attn_windows.view(-1, self.window_size, self.window_size, C)
shifted_x = self.window_reverse(attn_windows, self.window_size, H, W)
# Reverse cyclic shift
if self.shift_size > 0:
x = torch.roll(shifted_x, shifts=(self.shift_size, self.shift_size), dims=(1, 2))
else:
x = shifted_x
x = x.view(B, H * W, C)
# FFN
x = shortcut + self.drop_path(x)
x = x + self.drop_path(self.mlp(self.norm2(x)))
return x
def window_partition(self, x: torch.Tensor, window_size: int) -> torch.Tensor:
B, H, W, C = x.shape
x = x.view(B, H // window_size, window_size, W // window_size, window_size, C)
windows = x.permute(0, 1, 3, 2, 4, 5).contiguous()
windows = windows.view(-1, window_size, window_size, C)
return windows
def window_reverse(self, windows: torch.Tensor, window_size: int,
H: int, W: int) -> torch.Tensor:
B = int(windows.shape[0] / (H * W / window_size / window_size))
x = windows.view(B, H // window_size, W // window_size, window_size, window_size, -1)
x = x.permute(0, 1, 3, 2, 4, 5).contiguous().view(B, H, W, -1)
return x
class PatchEmbedding(nn.Module):
"""Patch embedding layer for Swin Transformer."""
def __init__(self, in_channels: int = 128, embed_dim: int = 64,
patch_size: int = 4):
super().__init__()
self.patch_embed = nn.Conv2d(in_channels, embed_dim,
kernel_size=patch_size, stride=patch_size)
def forward(self, x: torch.Tensor) -> Tuple[torch.Tensor, int, int]:
x = self.patch_embed(x)
B, C, H, W = x.shape
x = x.flatten(2).transpose(1, 2)
return x, H, W
class SwinTransformerStage(nn.Module):
"""Swin Transformer stage with multiple blocks."""
def __init__(self, dim: int, depth: int, num_heads: int = 8,
window_size: int = 4, drop_path: List[float] = None):
super().__init__()
self.blocks = nn.ModuleList([
SwinTransformerBlock(
dim=dim, num_heads=num_heads, window_size=window_size,
shift_size=0 if i % 2 == 0 else window_size // 2,
drop_path=drop_path[i] if drop_path else 0.
)
for i in range(depth)
])
def forward(self, x: torch.Tensor, H: int, W: int) -> torch.Tensor:
for block in self.blocks:
x = block(x, H, W)
return x
# ============================================================================
# EFFICIENTNETV2S WITH SWIN TRANSFORMER
# ============================================================================
class MBConv(nn.Module):
"""Mobile Inverted Bottleneck Conv."""
def __init__(self, in_channels: int, out_channels: int, expand_ratio: int,
kernel_size: int = 3, stride: int = 1, se_ratio: float = 0.25):
super().__init__()
self.use_residual = stride == 1 and in_channels == out_channels
hidden_dim = in_channels * expand_ratio
layers = []
# Expansion
if expand_ratio != 1:
layers.extend([
nn.Conv2d(in_channels, hidden_dim, 1, bias=False),
nn.BatchNorm2d(hidden_dim),
nn.SiLU(inplace=True)
])
# Depthwise
layers.extend([
nn.Conv2d(hidden_dim, hidden_dim, kernel_size, stride,
kernel_size//2, groups=hidden_dim, bias=False),
nn.BatchNorm2d(hidden_dim),
nn.SiLU(inplace=True)
])
# Squeeze-and-Excitation
if se_ratio > 0:
se_dim = max(1, int(in_channels * se_ratio))
layers.append(nn.Sequential(
nn.AdaptiveAvgPool2d(1),
nn.Conv2d(hidden_dim, se_dim, 1),
nn.SiLU(inplace=True),
nn.Conv2d(se_dim, hidden_dim, 1),
nn.Sigmoid()
))
# Output projection
layers.extend([
nn.Conv2d(hidden_dim, out_channels, 1, bias=False),
nn.BatchNorm2d(out_channels)
])
self.conv = nn.Sequential(*layers)
def forward(self, x: torch.Tensor) -> torch.Tensor:
if self.use_residual:
return x + self.conv(x)
return self.conv(x)
class FusedMBConv(nn.Module):
"""Fused Mobile Inverted Bottleneck Conv (EfficientNetV2)."""
def __init__(self, in_channels: int, out_channels: int, expand_ratio: int,
kernel_size: int = 3, stride: int = 1, se_ratio: float = 0.25):
super().__init__()
self.use_residual = stride == 1 and in_channels == out_channels
hidden_dim = in_channels * expand_ratio
layers = []
# Fused expansion + depthwise
layers.extend([
nn.Conv2d(in_channels, hidden_dim, kernel_size, stride,
kernel_size//2, bias=False),
nn.BatchNorm2d(hidden_dim),
nn.SiLU(inplace=True)
])
# Squeeze-and-Excitation
if se_ratio > 0 and expand_ratio != 1:
se_dim = max(1, int(in_channels * se_ratio))
layers.append(nn.Sequential(
nn.AdaptiveAvgPool2d(1),
nn.Conv2d(hidden_dim, se_dim, 1),
nn.SiLU(inplace=True),
nn.Conv2d(se_dim, hidden_dim, 1),
nn.Sigmoid()
))
# Output projection
if expand_ratio != 1:
layers.extend([
nn.Conv2d(hidden_dim, out_channels, 1, bias=False),
nn.BatchNorm2d(out_channels)
])
self.conv = nn.Sequential(*layers)
def forward(self, x: torch.Tensor) -> torch.Tensor:
if self.use_residual:
return x + self.conv(x)
return self.conv(x)
class EfficientNetV2S_Swin(nn.Module):
"""
EfficientNetV2S with embedded Swin Transformer at Stage 4.
"""
def __init__(self, num_classes: int = 8, pretrained: bool = True):
super().__init__()
# Stem
self.stem = nn.Sequential(
nn.Conv2d(3, 24, kernel_size=3, stride=2, padding=1, bias=False),
nn.BatchNorm2d(24),
nn.SiLU(inplace=True)
)
# Stage 1: Fused-MBConv1, depth=2
self.stage1 = self._make_fused_stage(24, 24, expand_ratio=1, depth=2)
# Stage 2: Fused-MBConv4, depth=4, stride=2
self.stage2 = self._make_fused_stage(24, 48, expand_ratio=4, depth=4, stride=2)
# Stage 3: Fused-MBConv4, depth=4
self.stage3 = self._make_fused_stage(48, 64, expand_ratio=4, depth=4)
# Stage 4: Parallel MBConv + Swin Transformer
self.stage4_conv = self._make_mbconv_stage(64, 128, expand_ratio=4, depth=1, stride=2)
# Swin Transformer branch
self.patch_embed = PatchEmbedding(in_channels=64, embed_dim=64, patch_size=4)
self.swin_stage = SwinTransformerStage(dim=64, depth=2, num_heads=8, window_size=4)
# Merge conv and swin outputs
self.stage4_merge = nn.Sequential(
nn.Conv2d(128 + 64, 128, 1, bias=False),
nn.BatchNorm2d(128),
nn.SiLU(inplace=True)
)
# Stage 5: MBConv6, depth=9
self.stage5 = self._make_mbconv_stage(128, 160, expand_ratio=6, depth=9)
# Stage 6: MBConv6, depth=15, stride=2
self.stage6 = self._make_mbconv_stage(160, 256, expand_ratio=6, depth=15, stride=2)
# Head
self.head = nn.Sequential(
nn.Conv2d(256, 1280, 1, bias=False),
nn.BatchNorm2d(1280),
nn.SiLU(inplace=True)
)
self.global_pool = nn.AdaptiveAvgPool2d(1)
self.feature_dim = 1280
# Initialize weights
self._initialize_weights()
def _make_fused_stage(self, in_c, out_c, expand_ratio, depth, stride=1):
layers = []
for i in range(depth):
s = stride if i == 0 else 1
in_ch = in_c if i == 0 else out_c
layers.append(FusedMBConv(in_ch, out_c, expand_ratio, stride=s))
return nn.Sequential(*layers)
def _make_mbconv_stage(self, in_c, out_c, expand_ratio, depth, stride=1):
layers = []
for i in range(depth):
s = stride if i == 0 else 1
in_ch = in_c if i == 0 else out_c
layers.append(MBConv(in_ch, out_c, expand_ratio, stride=s))
return nn.Sequential(*layers)
def _initialize_weights(self):
for m in self.modules():
if isinstance(m, nn.Conv2d):
nn.init.kaiming_normal_(m.weight, mode='fan_out', nonlinearity='relu')
elif isinstance(m, nn.BatchNorm2d):
nn.init.ones_(m.weight)
nn.init.zeros_(m.bias)
def forward(self, x: torch.Tensor) -> torch.Tensor:
x = self.stem(x)
x = self.stage1(x)
x = self.stage2(x)
x = self.stage3(x)
# Stage 4: Parallel processing
conv_branch = self.stage4_conv(x)
# Swin branch
swin_x, H, W = self.patch_embed(x)
swin_x = self.swin_stage(swin_x, H, W)
swin_x = swin_x.transpose(1, 2).view(x.size(0), 64, H, W)
swin_x = F.interpolate(swin_x, size=conv_branch.shape[2:],
mode='bilinear', align_corners=False)
# Merge
x = torch.cat([conv_branch, swin_x], dim=1)
x = self.stage4_merge(x)
x = self.stage5(x)
x = self.stage6(x)
x = self.head(x)
x = self.global_pool(x)
x = x.flatten(1)
return x
# ============================================================================
# COMPLETE TRANSXV2S-NET MODEL
# ============================================================================
class TransXV2SNet(nn.Module):
"""
Complete TransXV2S-Net: Hybrid ensemble of EfficientNetV2S+Swin and Modified Xception+DCGAN.
"""
def __init__(self, num_classes: int = 8, k: int = 64,
dropout: float = 0.3, pretrained: bool = False):
super().__init__()
self.num_classes = num_classes
# Branch 1: EfficientNetV2S + Swin Transformer
self.effnet_swin = EfficientNetV2S_Swin(num_classes=num_classes,
pretrained=pretrained)
effnet_dim = self.effnet_swin.feature_dim
# Branch 2: Modified Xception + DCGAN
self.xception_dcgab = ModifiedXception(num_classes=num_classes, k=k)
xception_dim = self.xception_dcgab.feature_dim
# Learnable fusion weights (alpha, beta for branch 1; gamma, delta for branch 2)
self.alpha = nn.Parameter(torch.tensor(0.5))
self.beta = nn.Parameter(torch.tensor(0.5))
self.gamma = nn.Parameter(torch.tensor(0.5))
self.delta = nn.Parameter(torch.tensor(0.5))
# Ensemble weights (theta, eta)
self.theta = nn.Parameter(torch.tensor(0.6))
self.eta = nn.Parameter(torch.tensor(0.4))
# Feature projection layers
self.effnet_proj = nn.Sequential(
nn.Linear(effnet_dim, 512),
nn.ReLU(inplace=True),
nn.Dropout(dropout)
)
self.xception_proj = nn.Sequential(
nn.Linear(xception_dim, 512),
nn.ReLU(inplace=True),
nn.Dropout(dropout)
)
# Final ensemble features
ensemble_dim = 1024 # 512 + 512
# Classification head
self.classifier = nn.Sequential(
nn.Linear(ensemble_dim, 512),
nn.ReLU(inplace=True),
nn.Dropout(dropout),
nn.Linear(512, num_classes)
)
self._initialize_weights()
def _initialize_weights(self):
for m in self.classifier.modules():
if isinstance(m, nn.Linear):
nn.init.kaiming_normal_(m.weight, mode='fan_out', nonlinearity='relu')
if m.bias is not None:
nn.init.constant_(m.bias, 0)
def forward(self, x: torch.Tensor) -> torch.Tensor:
# Branch 1: EfficientNetV2S + Swin
effnet_features = self.effnet_swin(x)
# Branch 2: Xception + DCGAN
xception_features = self.xception_dcgab(x)
# Project features
effnet_proj = self.effnet_proj(effnet_features)
xception_proj = self.xception_proj(xception_features)
# Ensemble fusion
# F_EffSwin = alpha * effnet_raw + beta * swin_component (implicit in forward)
# Here we use the learned projection as the combined representation
# Weighted ensemble
ensemble_features = torch.cat([self.theta * effnet_proj,
self.eta * xception_proj], dim=1)
# Classification
logits = self.classifier(ensemble_features)
return logits
def get_feature_maps(self, x: torch.Tensor) -> dict:
"""Get intermediate feature maps for visualization."""
# This would require modifying individual branches to return intermediates
pass
# ============================================================================
# TRAINING UTILITIES
# ============================================================================
class HairRemovalTransform:
"""
Hair removal preprocessing using morphological operations.
Simplified version - for production, use more sophisticated methods.
"""
def __init__(self, kernel_size: int = 9):
self.kernel_size = kernel_size
def __call__(self, img: torch.Tensor) -> torch.Tensor:
# Placeholder - implement actual hair removal
return img
class TransXV2SNetLoss(nn.Module):
"""
Custom loss with class balancing for imbalanced skin lesion datasets.
"""
def __init__(self, class_weights: Optional[torch.Tensor] = None,
label_smoothing: float = 0.1):
super().__init__()
self.ce = nn.CrossEntropyLoss(weight=class_weights,
label_smoothing=label_smoothing)
def forward(self, logits: torch.Tensor, targets: torch.Tensor) -> torch.Tensor:
return self.ce(logits, targets)
def create_optimizer(model: nn.Module, lr: float = 0.001, weight_decay: float = 1e-4):
"""Create Adamax optimizer as specified in paper."""
# Separate parameters for different learning rates
base_params = []
dcgab_params = []
for name, param in model.named_parameters():
if 'dcgab' in name or 'dcgan' in name:
dcgab_params.append(param)
else:
base_params.append(param)
optimizer = torch.optim.Adamax([
{'params': base_params, 'lr': lr},
{'params': dcgab_params, 'lr': lr * 0.1}
], weight_decay=weight_decay)
return optimizer
def get_lr_scheduler(optimizer: torch.optim.Optimizer, mode: str = 'plateau',
patience: int = 1, factor: float = 0.5):
"""Learning rate scheduler."""
if mode == 'plateau':
return torch.optim.lr_scheduler.ReduceLROnPlateau(
optimizer, mode='min', factor=factor, patience=patience, verbose=True
)
else:
return torch.optim.lr_scheduler.CosineAnnealingLR(optimizer, T_max=100)
# ============================================================================
# MODEL SUMMARY AND TESTING
# ============================================================================
def count_parameters(model: nn.Module) -> int:
"""Count trainable parameters."""
return sum(p.numel() for p in model.parameters() if p.requires_grad)
def test_model():
"""Test the complete model architecture."""
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
print(f"Testing TransXV2S-Net on {device}")
print("=" * 60)
# Create model
model = TransXV2SNet(num_classes=8, k=64, dropout=0.3)
model = model.to(device)
# Count parameters
total_params = count_parameters(model)
print(f"Total trainable parameters: {total_params:,} ({total_params/1e6:.2f}M)")
# Test forward pass
batch_size = 2
input_size = 128 # As specified in paper
x = torch.randn(batch_size, 3, input_size, input_size).to(device)
print(f"\nInput shape: {x.shape}")
with torch.no_grad():
output = model(x)
print(f"Output shape: {output.shape}")
print(f"Output range: [{output.min():.3f}, {output.max():.3f}]")
# Test individual components
print("\n" + "=" * 60)
print("Component Testing:")
print("=" * 60)
# Test DCGAN module
dcgab = DCGANModule(in_channels=256, k=64).to(device)
test_feat = torch.randn(1, 256, 16, 16).to(device)
dcgab_out = dcgab(test_feat)
print(f"DCGAN: {test_feat.shape} -> {dcgab_out.shape}")
# Test EfficientNetV2S+Swin
effnet = EfficientNetV2S_Swin(num_classes=8).to(device)
effnet_out = effnet(x)
print(f"EfficientNetV2S+Swin: {x.shape} -> {effnet_out.shape}")
# Test Modified Xception
xception = ModifiedXception(num_classes=8, k=64).to(device)
xception_out = xception(x)
print(f"Modified Xception: {x.shape} -> {xception_out.shape}")
print("\n" + "=" * 60)
print("All tests passed!")
print("=" * 60)
return model
# ============================================================================
# MAIN EXECUTION
# ============================================================================
if __name__ == "__main__":
# Run tests
model = test_model()
# Example training setup
print("\nExample training configuration:")
print(f" Optimizer: Adamax")
print(f" Initial LR: 0.001")
print(f" Batch size: 16")
print(f" Epochs: 25")
print(f" Input size: 128x128")
print(f" Early stopping patience: 3 epochs")References
Related posts, You May like to read
- 7 Shocking Truths About Knowledge Distillation: The Good, The Bad, and The Breakthrough (SAKD)
- 7 Revolutionary Breakthroughs in Medical Image Translation (And 1 Fatal Flaw That Could Derail Your AI Model)
- TimeDistill: Revolutionizing Time Series Forecasting with Cross-Architecture Knowledge Distillation
- HiPerformer: A New Benchmark in Medical Image Segmentation with Modular Hierarchical Fusion
- GeoSAM2 3D Part Segmentation — Prompt-Controllable, Geometry-Aware Masks for Precision 3D Editing
- DGRM: How Advanced AI is Learning to Detect Machine-Generated Text Across Different Domains
- A Knowledge Distillation-Based Approach to Enhance Transparency of Classifier Models
- Towards Trustworthy Breast Tumor Segmentation in Ultrasound Using AI Uncertainty
- Discrete Migratory Bird Optimizer with Deep Transfer Learning for Multi-Retinal Disease Detection
- How AI Combines Medical Images and Patient Data to Detect Skin Cancer More Accurately: A Deep Dive into Multimodal Deep Learning