Pneumonia remains a leading cause of global mortality, particularly among children and the elderly. Early detection is critical for improving survival rates, but traditional diagnostic methods rely heavily on chest X-rays (CXRs), which can be subjective and time-consuming for radiologists. Even subtle abnormalities in X-rays—such as lung opacities or fluid buildup—are often imperceptible to the human eye, leading to delayed diagnoses.
The World Health Organization (WHO) reports that pneumonia accounts for 15% of all deaths in children under five, emphasizing the urgent need for faster, more accurate diagnostic tools. Enter artificial intelligence (AI): Advanced machine learning and deep learning models are now transforming medical imaging, offering hope for rapid, automated pneumonia detection.
Current Methods in AI for Pneumonia Detection
Recent advancements in AI have introduced various approaches to analyze chest X-rays, including:
- Convolutional Neural Networks (CNNs): Pre-trained models like ResNet and MobileNetV2 extract hierarchical features from raw images.
- Feature Engineering: Techniques like Gray Level Co-Occurrence Matrix (GLCM) and wavelet transforms highlight texture patterns and spatial relationships in X-rays.
- Ensemble Models: Combining multiple algorithms to improve robustness and accuracy.
However, standalone models often face limitations:
- Class Imbalance: Datasets may have more pneumonia cases than normal ones, skewing results.
- Limited Generalizability: Models trained on specific datasets (e.g., adult patients) may fail on pediatric X-rays.
- Computational Complexity: Deep learning models require significant resources for training and inference.
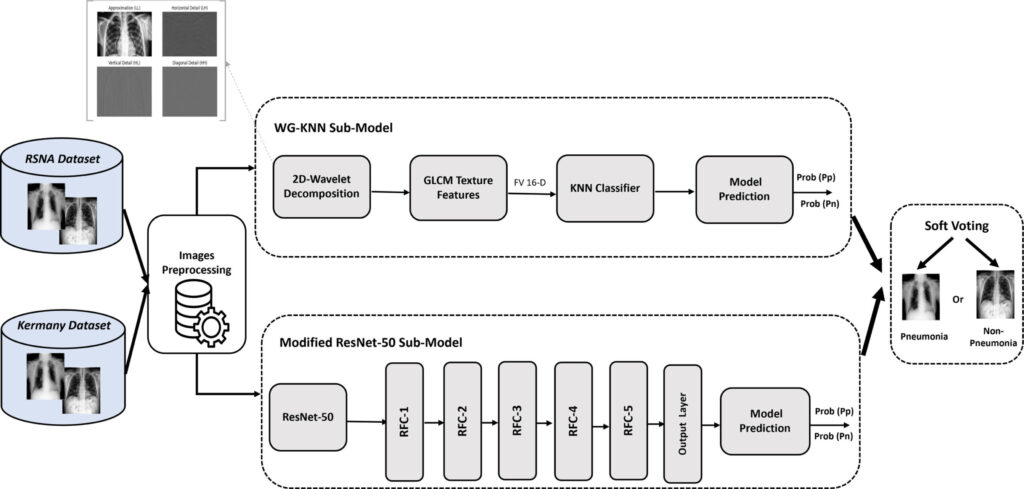
The Novel Fusion Model: Combining Strengths of KNN and ResNet-50
A groundbreaking study published in Intelligent Systems with Applications introduces Res-WG-KNN, a fusion model that marries engineered features with deep learning to achieve unprecedented accuracy in pneumonia detection. Here’s how it works:
Key Components of the Res-WG-KNN Model
- Wavelet Transforms and GLCM Texture Analysis
- 2D Discrete Wavelet Transform (DWT): Breaks down X-rays into frequency sub-bands, capturing multi-scale details like lung textures and edges.
- GLCM Features: Analyzes spatial pixel relationships to quantify texture properties such as contrast, energy, and homogeneity.
- Why it matters: These features reveal patterns invisible to the naked eye, such as early-stage inflammation.
- Deep Learning with Modified ResNet-50
- A pre-trained ResNet-50 model is fine-tuned to extract high-level features from raw X-rays.
- Five regularized fully connected (RFC) layers reduce dimensionality while preventing overfitting.
- Soft Voting Ensemble Strategy
- Combines predictions from the KNN classifier (using wavelet-GLCM features) and ResNet-50.
- Instead of a majority vote, it averages probabilities, ensuring balanced contributions from both models.
If you’re interested in solving classification problems, you may also find this article helpful: CACTUS Framework: Revolutionizing Cardiac Care with Deep Transfer Learning in Ultrasound Imaging
Breakthrough Results: Outperforming State-of-the-Art Models
The Res-WG-KNN model was tested on two public datasets:
- RSNA Dataset (Adult Patients): 30,227 DICOM images.
- Kermany Dataset (Pediatric Patients): 5,856 JPEG images.
Performance Highlights
| Metric | RSNA Dataset | Kermany Dataset |
|---|---|---|
| Accuracy | 97.0% | 99.0% |
| Precision | 97.5% | 98.3% |
| AUC Score | 0.97 | 0.99 |
The fusion model outperformed existing approaches like standalone ResNet-50, MobileNetV2, and ensemble models by 7–15% in accuracy, demonstrating its ability to generalize across diverse datasets.
Why This Matters for Healthcare
- Early Detection Saves Lives: Faster diagnosis enables timely treatment, reducing complications.
- Reduced Workload for Radiologists: AI acts as a “second opinion,” prioritizing urgent cases.
- Cost-Effective Scaling: The model’s efficiency makes it viable for low-resource settings.
Future Applications
- COVID-19 and Tuberculosis Detection: Adapting the model to identify other lung diseases.
- Integration with EHR Systems: Real-time analysis within hospital workflows.
- Global Health Equity: Deploying AI tools in underserved regions with limited radiologists.
FAQ: Addressing Common Questions
Q: How does the fusion model handle different X-ray formats (DICOM vs. JPEG)?
A: The model standardizes images to 224×224 pixels, ensuring compatibility across formats.
Q: Can it distinguish between viral and bacterial pneumonia?
A: Not yet—this requires additional clinical data, but future updates could incorporate lab results.
Q: Is the model biased toward adult or pediatric cases?
A: No—it achieved high accuracy on both datasets, proving its versatility.
Conclusion and Call-to-Action
The Res-WG-KNN model represents a leap forward in AI-driven pneumonia detection. By combining wavelet transforms, texture analysis, and deep learning, it addresses critical gaps in current diagnostic practices.
Ready to Explore AI in Healthcare?
- For Hospitals: Integrate cutting-edge AI tools to enhance diagnostic accuracy.
- For Researchers: Collaborate on advancing models for other respiratory diseases.
- For Developers: Access open-source datasets to build your own solutions.
Join the revolution in medical imaging—contact us today to learn how AI can transform your diagnostic workflows.
Based on the detailed information provided in the paper, I will reconstruct the complete code for the proposed methodology.
# Import required libraries
import numpy as np
import pandas as pd
import tensorflow as tf
from tensorflow.keras.applications import ResNet50
from tensorflow.keras.layers import Dense, Dropout, BatchNormalization
from tensorflow.keras.models import Model
from tensorflow.keras.optimizers import Adam
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import accuracy_score, roc_auc_score
from skimage.feature import greycomatrix, greycoprops
import pywt # For wavelet transforms
import pydicom # For DICOM processing
from PIL import Image
# ----------------------------
# Step 1: Data Loading & Preprocessing
# ----------------------------
def load_and_preprocess_data(image_paths, labels, target_size=(224, 224)):
images = []
for path in image_paths:
if path.endswith('.dcm'): # DICOM (RSNA dataset)
dicom = pydicom.dcmread(path)
img = dicom.pixel_array
else: # JPEG (Kermany dataset)
img = Image.open(path).convert('L')
img = img.resize(target_size)
img = np.array(img) / 255.0 # Normalize
images.append(img)
return np.array(images), np.array(labels)
# Example usage:
# X_train, y_train = load_and_preprocess(train_paths, train_labels)
# ----------------------------
# Step 2: Feature Extraction (WG-KNN Sub-model)
# ----------------------------
def extract_wavelet_glcm_features(image, wavelet='haar'):
# Apply 2D Discrete Wavelet Transform (DWT)
coeffs = pywt.dwt2(image, wavelet)
LL, (LH, HL, HH) = coeffs
# Compute GLCM for each sub-band
features = []
for sub_band in [LL, LH, HL, HH]:
glcm = greycomatrix(sub_band.astype(np.uint8), distances=[1], angles=[0], symmetric=True, normed=True)
contrast = greycoprops(glcm, 'contrast')[0, 0]
energy = greycoprops(glcm, 'energy')[0, 0]
homogeneity = greycoprops(glcm, 'homogeneity')[0, 0]
dissimilarity = greycoprops(glcm, 'dissimilarity')[0, 0]
features.extend([contrast, energy, homogeneity, dissimilarity])
return np.array(features)
# Extract features for all images
def extract_features_batch(images):
features = []
for img in images:
features.append(extract_wavelet_glcm_features(img))
return np.array(features)
# ----------------------------
# Step 3: Train KNN Classifier
# ----------------------------
def train_knn(X_train_features, y_train):
param_grid = {
'n_neighbors': [25, 50, 75],
'weights': ['uniform', 'distance'],
'algorithm': ['auto']
}
knn = KNeighborsClassifier()
grid_search = GridSearchCV(knn, param_grid, cv=5, scoring='accuracy')
grid_search.fit(X_train_features, y_train)
best_knn = grid_search.best_estimator_
return best_knn
# ----------------------------
# Step 4: Modified ResNet-50 Sub-model
# ----------------------------
def build_resnet50_model(input_shape=(224, 224, 3), num_classes=1):
base_model = ResNet50(weights='imagenet', include_top=False, input_shape=input_shape)
base_model.trainable = False # Freeze ResNet layers
x = base_model.output
x = tf.keras.layers.GlobalAveragePooling2D()(x)
# Add 5 RFC blocks
for units in [512, 256, 128, 64, 32]:
x = Dense(units)(x)
x = BatchNormalization()(x)
x = Dropout(0.3)(x)
x = tf.keras.activations.relu(x)
predictions = Dense(num_classes, activation='sigmoid')(x)
model = Model(inputs=base_model.input, outputs=predictions)
model.compile(
optimizer=Adam(learning_rate=0.01),
loss='binary_crossentropy',
metrics=['accuracy']
)
return model
# ----------------------------
# Step 5: Soft Voting Ensemble
# ----------------------------
def soft_voting(knn_probs, resnet_probs):
avg_probs = (knn_probs + resnet_probs) / 2
return np.argmax(avg_probs, axis=1)
# ----------------------------
# Step 6: Full Pipeline
# ----------------------------
def main():
# Load datasets (Replace with your paths)
# RSNA (DICOM) and Kermany (JPEG) datasets
X_train, y_train = load_and_preprocess(train_paths, train_labels)
X_test, y_test = load_and_preprocess(test_paths, test_labels)
# --- WG-KNN Sub-model ---
# Extract engineered features
X_train_features = extract_features_batch(X_train)
X_test_features = extract_features_batch(X_test)
# Train KNN
knn_model = train_knn(X_train_features, y_train)
knn_probs = knn_model.predict_proba(X_test_features)
# --- ResNet-50 Sub-model ---
# Convert grayscale to RGB for ResNet input
X_train_rgb = np.stack([X_train]*3, axis=-1)
X_test_rgb = np.stack([X_test]*3, axis=-1)
resnet_model = build_resnet50_model()
resnet_model.fit(X_train_rgb, y_train, epochs=100, batch_size=256, validation_split=0.1)
resnet_probs = resnet_model.predict(X_test_rgb)
# --- Ensemble Predictions ---
final_preds = soft_voting(knn_probs, resnet_probs)
# Evaluate
accuracy = accuracy_score(y_test, final_preds)
auc = roc_auc_score(y_test, final_preds)
print(f"Accuracy: {accuracy:.2%}, AUC: {auc:.2f}")
if __name__ == "__main__":
main()