In today’s fast-paced medical landscape, early detection of skin cancer is more crucial than ever. With skin cancer cases on the rise due to increased ultraviolet exposure and environmental factors, accurate and efficient diagnostic tools are essential. Enter SILP – a novel system that leverages state-of-the-art machine learning techniques to enhance skin lesion classification. In this article, we explore the groundbreaking SILP model, its underlying technology, and how it is revolutionizing skin cancer detection.
Introduction to Skin Lesion Classification
Skin lesions come in many forms and can be challenging to diagnose accurately. Traditional methods rely on visual examination by dermatologists and histopathological analysis, which, while effective, are subject to human error and variability. Modern deep learning techniques are now making a significant impact in the medical field by providing automated, precise, and accessible diagnostic tools.
Key keywords:
- Skin lesion classification
- Skin cancer detection
- Dermatology diagnostics
What is SILP?
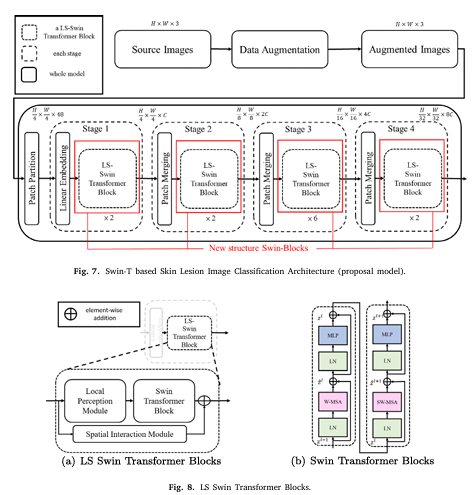
SILP stands for Spatial Interaction and Local Perception, and it represents a new frontier in skin lesion classification. By integrating advanced machine learning architectures with innovative modules that capture both local details and global spatial relationships, SILP offers superior performance compared to conventional models.
The Science Behind SILP
The SILP model is built on the robust Swin Transformer architecture, which has already proven its effectiveness in image classification tasks. However, SILP goes further by incorporating two critical components:
- Local Perception Module (LPM): Enhances feature extraction by capturing fine-grained, local details in images.
- Spatial Interaction Module (SIM): Improves the model’s ability to understand global spatial relationships between pixels, overcoming limitations of the fixed-window approach inherent in many transformer models.
How SILP Works: A Detailed Overview
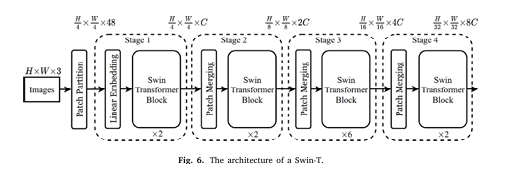
1. Swin Transformer Backbone
At the core of SILP is the Swin Transformer (Swin-T), a hierarchical vision transformer known for its efficiency and performance in image recognition tasks. Unlike traditional convolutional neural networks (CNNs), Swin Transformer processes images using a shifted window mechanism. This method divides images into smaller patches and processes them hierarchically, preserving spatial information while reducing computational overhead.
Benefits:
2. Local Perception Module (LPM)
The LPM is specifically designed to tackle one of the most critical challenges in skin lesion classification: the extraction of meaningful features from images with high variability. Skin lesions often vary widely in shape, color, and size. The LPM uses dilated convolutions to expand the receptive field without losing resolution, allowing the model to capture subtle textural and structural details.

Key advantages of LPM:
3. Spatial Interaction Module (SIM)
While the Swin Transformer efficiently captures local patterns within defined windows, it can miss relationships between these windows. The Spatial Interaction Module (SIM) addresses this gap by incorporating an attention mechanism that enables cross-window communication. This module ensures that the model understands the broader spatial context, which is essential for accurate skin lesion detection.

SIM highlights:
5. Activation Function Optimization: SiLU vs. GELU
Training stability and efficiency are critical in deep learning. SILP replaces the Gaussian Error Linear Unit (GELU) with the Sigmoid Linear Unit (SiLU) activation function. The SiLU function is computationally simpler and has been shown to propagate gradients more effectively, leading to faster training convergence and improved overall performance.
Why SiLU matters:
The Impact of SILP on Skin Cancer Detection
The SILP model has undergone rigorous testing on two widely recognized skin lesion datasets: HAM10000 and ISIC2017. Experimental results have demonstrated that SILP outperforms current state-of-the-art models in several key evaluation metrics:
| MODEL | ACCURACY | SENSITIVITY | SPECIFICITY |
| Swin-T (Baseline) | 95.2% | 93.2% | 91.4% |
| DPE-BoTNet | 95.8% | 90.1% | 91.9% |
| SILP (Proposed) | 96.9% | 95.0% | 93.2% |
Addressing the Data Imbalance Challenge
Medical image datasets often suffer from class imbalance, where some lesion types are underrepresented. SILP mitigates this issue through advanced data augmentation techniques, including Mixup and grayscale conversion, which not only increase dataset diversity but also improve model generalization. This robust handling of imbalanced data is key to ensuring reliable skin cancer detection across different populations.
Benefits for Medical Practitioners and Patients
For Dermatologists
- Enhanced Diagnostic Confidence: With higher accuracy and reliable sensitivity, dermatologists can confidently use SILP as an auxiliary tool in their diagnostic process.
- Reduced Workload: Automated image classification allows practitioners to focus on complex cases and reduce the time spent on routine examinations.
- Improved Patient Outcomes: Early and accurate detection of skin cancer can lead to timely treatment, ultimately saving lives.
For Patients
- Accessible Screening: SILP-based applications can be integrated into telemedicine platforms, making skin cancer screening accessible even in remote areas.
- Quick Results: Rapid and reliable diagnostic support helps patients receive early interventions, improving overall treatment efficacy.
- Cost-effective Care: Reducing the need for invasive procedures and multiple consultations can lower healthcare costs for patients.
Conclusion
In the realm of skin cancer detection, early and accurate diagnosis is paramount. SILP’s innovative approach, which leverages the Swin Transformer along with Local Perception and Spatial Interaction modules, marks a significant advancement in skin lesion classification. This model not only sets a new standard in accuracy and performance but also promises to transform the way dermatological diagnoses are conducted, ultimately contributing to improved patient care.
Are you ready to harness the power of AI in healthcare?
Discover how the SILP model can revolutionize your diagnostic practices. Whether you’re a healthcare professional looking to enhance diagnostic accuracy or a tech enthusiast interested in the latest in medical AI, SILP offers a glimpse into the future of skin cancer detection. Get in touch today to learn more about implementing SILP in your practice and join the revolution in AI-driven medical diagnostics.
Here, Complete implementation code of SILP in Pytorch.
import torch
import torch.nn as nn
from timm.models.swin_transformer import SwinTransformerBlock, PatchEmbed
# Custom Activation: SiLU (replaces GELU)
class SiLU(nn.Module):
def forward(self, x):
return x * torch.sigmoid(x)
# Local Perception Module (LPM)
class LocalPerceptionModule(nn.Module):
def __init__(self, in_channels, dilation=2):
super().__init__()
self.dilated_conv = nn.Conv2d(
in_channels, in_channels, kernel_size=3,
padding=dilation, dilation=dilation, groups=in_channels
)
self.act = nn.GELU() # GELU used in LPM per paper
def forward(self, x):
B, L, C = x.shape # (Batch, Sequence Length, Channels)
H = W = int(L ** 0.5)
x = x.view(B, H, W, C).permute(0, 3, 1, 2) # Reshape to spatial
x = self.dilated_conv(x)
x = self.act(x)
x = x.permute(0, 2, 3, 1).view(B, L, C) # Reshape back
return x
# Spatial Interaction Module (SIM)
class SpatialInteractionModule(nn.Module):
def __init__(self, in_channels):
super().__init__()
self.reduce = nn.Conv2d(in_channels, in_channels // 2, kernel_size=1)
self.norm = nn.LayerNorm(in_channels // 2)
self.act = nn.GELU()
def forward(self, x):
B, L, C = x.shape
H = W = int(L ** 0.5)
x = x.view(B, H, W, C).permute(0, 3, 1, 2)
x = self.reduce(x) # Reduce channels
x = x.permute(0, 2, 3, 1).view(B, L, -1)
x = self.norm(x)
x = self.act(x)
# Average pooling in vertical/horizontal directions
x_spatial = x.view(B, H, W, -1)
v_h = torch.mean(x_spatial, dim=2) # (B, H, C/2)
v_w = torch.mean(x_spatial, dim=1) # (B, W, C/2)
attention = torch.einsum('bhc,bwc->bhwc', v_h, v_w).view(B, H*W, -1)
return attention
# Modified Swin-T Block with LPM and SIM
class LS_SwinBlock(nn.Module):
def __init__(self, dim, input_resolution, num_heads, window_size=7, shift_size=0):
super().__init__()
# Original Swin-T components
self.swin_block = SwinTransformerBlock(
dim=dim,
input_resolution=input_resolution,
num_heads=num_heads,
window_size=(window_size, window_size), # Pass as tuple
shift_size=(shift_size, shift_size) if shift_size > 0 else (0, 0)
)
# Custom modules
self.lpm = LocalPerceptionModule(dim)
self.sim = SpatialInteractionModule(dim)
# Modified MLP with SiLU
self.mlp = nn.Sequential(
nn.Linear(dim, 4 * dim),
SiLU(),
nn.Dropout(0.1),
nn.Linear(4 * dim, dim),
nn.Dropout(0.1)
)
def forward(self, x):
# Path 1: LPM + Swin-T Block
x1 = self.lpm(x)
x1 = self.swin_block(x1)
# Path 2: SIM
x2 = self.sim(x)
# Combine paths
x = x1 + x2
# MLP
x = self.mlp(x)
return x
# Full SILP Model
class SILP(nn.Module):
def __init__(self, num_classes=7, embed_dim=96, img_size=224, window_size=7):
super().__init__()
# Patch Embedding (Swin-T backbone)
self.patch_embed = PatchEmbed(img_size=img_size, patch_size=4, in_chans=3, embed_dim=embed_dim)
self.input_resolution = self.patch_embed.grid_size # (H, W)
# Stage 1-4 with LS-Swin Blocks
self.stages = nn.ModuleList([
self._make_stage(embed_dim, self.input_resolution, num_heads=3, num_blocks=2, shift_size=0),
self._make_stage(embed_dim*2, (self.input_resolution[0]//2, self.input_resolution[1]//2),
num_heads=6, num_blocks=2, shift_size=window_size//2),
self._make_stage(embed_dim*4, (self.input_resolution[0]//4, self.input_resolution[1]//4),
num_heads=12, num_blocks=6, shift_size=0),
self._make_stage(embed_dim*8, (self.input_resolution[0]//8, self.input_resolution[1]//8),
num_heads=24, num_blocks=2, shift_size=window_size//2),
])
# Classifier
self.avgpool = nn.AdaptiveAvgPool1d(1)
self.fc = nn.Linear(embed_dim*8, num_classes)
def _make_stage(self, dim, input_resolution, num_heads, num_blocks, shift_size=0):
blocks = []
for _ in range(num_blocks):
blocks.append(
LS_SwinBlock(
dim=dim,
input_resolution=input_resolution,
num_heads=num_heads,
window_size=7,
shift_size=shift_size if _ % 2 == 0 else 0 # Alternate shifted/non-shifted
)
)
return nn.Sequential(*blocks)
def forward(self, x):
x = self.patch_embed(x) # (B, L, C)
for stage in self.stages:
x = stage(x)
x = self.avgpool(x.transpose(1, 2)).squeeze()
x = self.fc(x)
return x
# Example Usage
model = SILP(num_classes=7)
print(model)Sources:
- Nguyen, K. et al. (2024). SILP: Enhancing Skin Lesion Classification. Expert Systems With Applications.
- Tschandl, P. et al. (2018). The HAM10000 Dataset. Scientific Data.
- American Academy of Dermatology (2023). Skin Cancer Incidence Report.
- Liu, Z., et al. (2021). Swin Transformer: Hierarchical Vision Transformer Using Shifted Windows. ICCV.

Pingback: Revolutionizing Brain Tumor Classification with AI Accuracy