Introduction
Imagine a radiologist spending hours manually tracing the intricate network of airways in thousands of patient CT scans, a tedious process prone to human error and fatigue. This reality affects millions of patients worldwide waiting for accurate diagnoses of life-threatening lung conditions. However, a groundbreaking advancement in artificial intelligence is poised to transform this landscape entirely.
Researchers at Imperial College London have developed DMGSA (Dynamical Multi-order Responses and Global Semantic-infused Adversarial network), a sophisticated deep learning model that automates airway segmentation in chest CT images with unprecedented accuracy. Published in Medical Image Analysis in 2025, this innovation addresses critical challenges that have plagued medical imaging for decades, particularly for patients with COVID-19, pulmonary fibrosis, and lung cancer.
The problem is urgent: Early detection of progressive fibrotic lung disease could save countless lives by enabling clinicians to initiate therapies before irreversible damage occurs. Yet manual visual assessment of airways introduces significant interobserver variability, poor reproducibility, and insensitivity to subtle changes. This article explores how DMGSA overcomes these barriers through four innovative technological components that fundamentally reimagine how machines learn from medical images.
Why Airway Segmentation Matters in Modern Medicine
The Critical Role of Airways in Diagnosis
The tracheobronchial tree—the network of air passages branching throughout the lungs—serves as a crucial diagnostic indicator for multiple life-threatening conditions. Traction bronchiectasis, the abnormal dilation of airways caused by surrounding lung fibrosis, has emerged as a robust predictor of outcomes in patients with pulmonary fibrosis. However, visual assessment alone is insufficient for modern precision medicine.
Consider the statistics: Pulmonary fibrosis accounts for approximately 1% of all deaths in the United Kingdom alone, with similar proportions reported globally. Without reliable baseline measurements, clinicians cannot accurately predict disease progression or determine optimal treatment timing. This gap between clinical need and diagnostic capability has driven researchers to develop automated solutions.
Current Challenges in Airway Segmentation
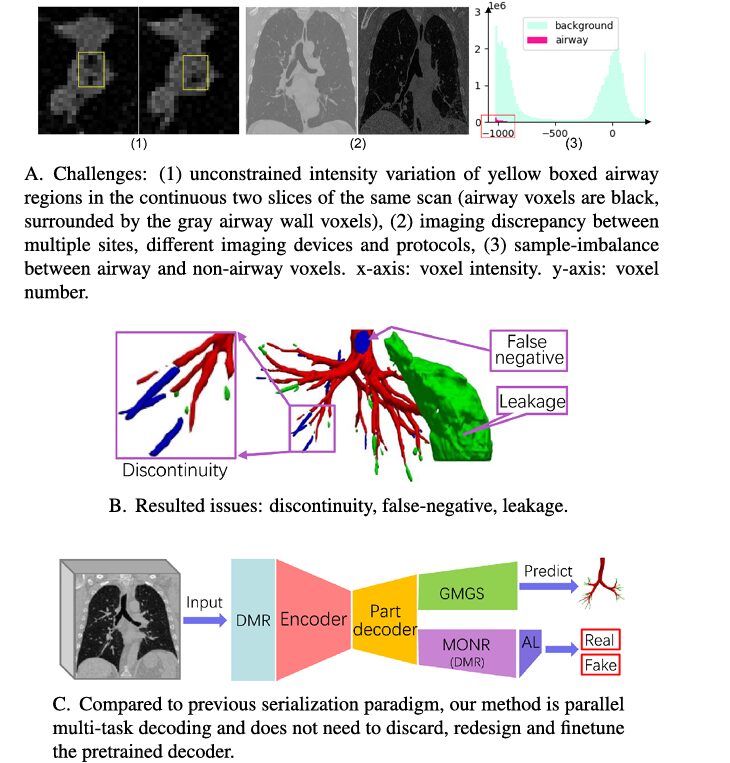
Traditional machine learning approaches struggle with several inherent limitations:
- Extreme class imbalance: Airways comprise only a small fraction of total lung volume, forcing models to distinguish tiny structures from massive amounts of background tissue
- Structural complexity: The airway tree contains thousands of branches ranging from thick central airways to ultrathin terminal bronchioles measuring mere millimeters
- Discontinuity and false negatives: Existing methods frequently miss thin bronchioles or predict fragmented rather than continuous airways
- Dataset scarcity: Expert annotations are extraordinarily labor-intensive, limiting available training data
- Unconstrained intensities: Variable CT scanning protocols produce unpredictable intensity patterns in airway tissues
These challenges have constrained even state-of-the-art methods to detect only 74-90% of bronchiole segments in benchmark datasets.
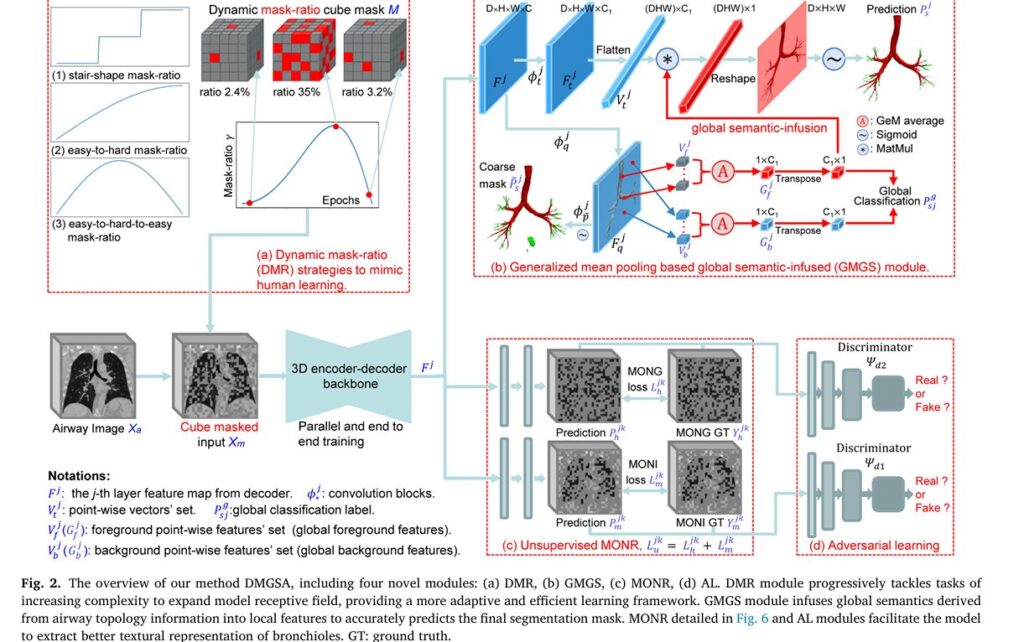
unsupervised and supervised learning, including (a) Dynamic Mask-Ratio (DMR)
module to mimic human learning, (b) Generalized Mean pooling based Global
Semantic-infused (GMGS) module to segment airway, (c) unsupervised Multi
Order Normalized Responses (MONR) and (d) Adversarial Learning (AL) mod
ules to learn abundant textures of bronchioles, aiding the supervised segmenta
tion branch in parallel.
The DMGSA Solution: Four Innovative Components
1. Dynamic Mask-Ratio (DMR): Learning Like Humans Do
Traditional deep learning methods train models with fixed masking percentages—essentially asking AI to solve equally difficult problems from day one. DMGSA introduces a paradigm shift inspired by human cognitive development.
The Dynamic Mask-Ratio principle implements a schedule where masking difficulty progressively increases throughout training, following the equation:
$$\gamma = \sin\left(\left(\frac{e_i}{e_{max}}\right)^{\rho} \cdot \pi\right) \cdot r, \quad \rho \in [1.0, 3.0], \quad r \in [0.15, 0.5]$$
Where:
- γ is the dynamic mask ratio
- ei represents the current training epoch
- emax is the total number of epochs
- ρ controls the speed of difficulty progression
- r sets the maximum masking ratio
This “easy-to-hard-to-easy” strategy enables the model to gradually expand its receptive field and learn increasingly complex long-range topology information about airways. Experimental results demonstrate a remarkable 1.12% improvement in comprehensive segmentation metrics (CCF score) compared to fixed-mask approaches.
Why this matters: Rather than learning from artificially masked data then discarding that capability at test time, DMGSA bridges the train-test gap by reducing mask ratios near training completion, creating a seamless transition to full input processing.
2. Multi-Order Normalized Responses (MONR): Capturing Hidden Texture
Airways contain abundant texture features—the criss-cross branching patterns and wall structures—that reveal diagnostic information invisible to standard image processing. DMGSA’s MONR module extracts this information through mathematical transformations that highlight different frequency components of medical images.
The process unfolds in three stages:
Stage 1 – Normalized Image Preparation: First, raw CT images are normalized to a standard intensity range [0, 255]:
$$X_a = \Phi_n(X) = \frac{X – \min(X)}{\max(X) – \min(X)} \cdot 255$$
Stage 2 – Multi-Order Exponential Normalization: Different exponential powers reveal distinct frequency information. The model generates Multi-Order Normalized Images (MONI) using orders k ∈ {0.5, 1.5}:
$$\tilde{X}_m^k = (X_a)^k$$ $$X_m^k = \left(\frac{\tilde{X}_m^k – \min(\tilde{X}_m^k)}{\max(\tilde{X}_m^k) – \min(\tilde{X}_m^k)}\right)^{1/k}$$
Stage 3 – Gradient Extraction: Using 2D Sobel operators applied across depth, width, and height dimensions, the model extracts Multi-Order Normalized orientation Gradients (MONG):
$$H_d^k = \Phi_d(\Phi_n(\tilde{X}_m^k), \text{Sobel_kernel}(d)), \quad d \in {\text{depth, width, height}}$$
The results are striking: The MONR module improved detection length ratio (DLR) by 2.17% and detected branch ratio (DBR) by 3.31% on fibrosis datasets—specifically enhancing segmentation of thin, difficult-to-detect terminal bronchioles. This targeted improvement directly addresses the false-negative problem that has plagued previous methods.
3. Generalized Mean Pooling Based Global Semantic-infused (GMGS) Module
Rather than processing each pixel location independently, the GMGS module infuses global understanding of airway topology into local feature extraction. This innovation draws from modern image retrieval techniques, adapting the concept for medical segmentation.
The module leverages trainable Generalized Mean (GeM) pooling:
$$G_x^j = \left(\frac{1}{|V_x^j|}\sum(V_x^j + V_{pe})^{\alpha}\right)^{1/\alpha}W + b_1$$
Where:
- Vx represents point-wise feature vectors
- α is a learnable power index (converged to 2.83 in experiments)
- Vpe adds position embedding to preserve topology information
The insight: Foreground airway features naturally cluster together in high-dimensional space, making their cosine similarities much larger than similarities between airway and background features. By maximizing this distinction through GeM pooling, the module learns to discriminate airways from surrounding tissue far more effectively.
Quantitative impact: The GMGS module reduced the airway missing ratio (AMR) by 0.47% while improving CCF scores by 0.95%—a substantial improvement for detecting structures that previous methods frequently missed entirely.
4. Adversarial Learning: Teaching Machines to Distinguish Real from Fake
Inspired by Generative Adversarial Networks (GANs), DMGSA employs two lightweight discriminators that compete with the segmentation network. This adversarial framework forces the model to generate increasingly realistic Multi-Order Normalized Response reconstructions.
Two discriminators operate in parallel:
- Ψ_{d1} evaluates Multi-Order Normalized Images (MONI) authenticity
- Ψ_{d2} evaluates Multi-Order Normalized Gradients (MONG) authenticity
The optimization plays out as a minimax game:
$$\min_{\theta_{pa}}\max_{\theta_{d1},\theta_{d2}} L(\theta_{pa}, \theta_{d1}, \theta_{d2}) = L_{pa}(X_m; \theta_{pa}) – \alpha L_{d1} – \beta L_{d2}$$
During each training step, discriminators attempt to correctly classify predicted reconstructions as “fake” while the segmentation network tries to fool them. This competition drives the network to extract increasingly subtle textural features of terminal bronchioles.
Experimental validation: While adversarial learning contributed a modest 0.39% improvement in CCF scores independently, its effects compound with other components. Visualizations clearly show enhanced focus on terminal bronchiole textures, directly addressing the false-negative problem in challenging anatomical regions.
Revolutionary Results: Performance Across Diverse Lung Pathologies
DMGSA was evaluated on three increasingly challenging datasets representing real-world clinical scenarios:
| Dataset | IoU (%) | DLR (%) | DBR (%) | CCF (%) | Improvement vs. Previous SOTA |
|---|---|---|---|---|---|
| BAS (Normal/Cancer) | 88.02 | 96.04 | 95.11 | 91.35 | +1.66% |
| COVID-19 Scans | 93.31 | 97.49 | 97.01 | 95.13 | +2.43% |
| Pulmonary Fibrosis | 84.01 | 87.12 | 82.17 | 85.28 | +4.29% |
Key takeaway: DMGSA’s most dramatic improvements emerge on fibrotic lung datasets—precisely where clinical need is greatest. The 4.29% improvement in comprehensive segmentation metrics on fibrosis cases represents a significant advance for a disease affecting millions worldwide.
Ablation Studies Confirm Component Synergy
The research team systematically disabled each module to quantify individual contributions:
- DMR alone: +1.12% CCF improvement through better receptive field modeling
- GMGS addition: +0.95% CCF improvement through global semantic infusion
- MONR integration: +1.02% CCF improvement through texture feature extraction
- Adversarial learning: +0.39% CCF improvement with compounding texture benefits
Critically, combining all components delivered cumulative 4.15% total improvement—exceeding the sum of individual contributions, demonstrating genuine synergy between innovations.
Clinical Implications and Real-World Applications
Early Disease Detection and Monitoring
DMGSA enables automated quantification of traction bronchiectasis severity—a proven predictor of pulmonary fibrosis progression. Radiologists can now obtain objective baseline measurements and track subtle changes over weeks or months, guiding treatment decisions before irreversible lung damage occurs.
Resource Efficiency and Accessibility
Manual airway segmentation requires 30-60 minutes per case for experienced radiologists. DMGSA processes cases in seconds, dramatically improving diagnostic throughput and making advanced analysis accessible to hospitals lacking specialized lung imaging expertise.
Reduced Interobserver Variability
Visual assessment introduces 15-25% variability between radiologists—a critical limitation for longitudinal studies and clinical trials. Automated segmentation provides objective, reproducible measurements essential for precision medicine.
Comparison with Existing State-of-the-Art Methods
How DMGSA Outperforms Previous Approaches
| Method | Year | Architecture | Key Innovation | DBR on Fibrosis |
|---|---|---|---|---|
| V-Net | 2016 | 3D CNN | Basic volumetric segmentation | 3.40% |
| WingNet | 2021 | 3D CNN + Attention | Gradient-based attention | 63.01% |
| NaviAirway | 2022 | 3D CNN | Bronchiole-sensitive loss | 51.47% |
| FANN (Previous SOTA) | 2023 | 3D CNN + Fuzzy Attention | Fuzzy logic integration | 73.44% |
| DMGSA (New) | 2025 | 3D CNN + Dynamic Masking + Adversarial | Multi-component synergy | 82.17% |
DMGSA’s 8.73 percentage point improvement over the previous state-of-the-art on fibrosis datasets represents a paradigm shift in capability.
Technical Innovation: Beyond Traditional Deep Learning
The Parallel Learning Paradigm
Unlike previous methods following a “pretrain-discard-finetune” paradigm, DMGSA trains unsupervised and supervised branches in parallel. This eliminates computational waste and reduces overfitting—particularly critical when working with limited medical imaging datasets.
Label Scarcity as a Design Feature
Rather than treating limited annotations as a limitation, DMGSA leverages unsupervised learning to extract maximum information from unlabeled CT volumes. This design philosophy makes the method particularly valuable for rare diseases and specialized applications where expert annotations are unavailable.
Future Directions and Broader Impact
Expanding to Other Organ Systems
The DMGSA architecture’s principles—dynamic masking, multi-order response prediction, and adversarial refinement—generalize beyond airways. Similar approaches could revolutionize blood vessel segmentation, tumor boundary detection, and skeletal structure analysis.
Integration with Clinical Workflows
Ongoing work focuses on integrating DMGSA predictions into radiology reporting systems, enabling semi-automated report generation and quantitative disease indices that improve prognostication.
Addressing Healthcare Disparities
By automating expert-level diagnostics, DMGSA can democratize advanced medical imaging analysis across healthcare systems with varying levels of specialized expertise, potentially reducing diagnostic disparities.
Conclusion: A New Standard in Medical Image Analysis
DMGSA represents a watershed moment in automated medical imaging. By synthesizing four distinct innovations—dynamic learning schedules mimicking human development, multi-order texture analysis extracting hidden diagnostic information, global semantic infusion connecting local details to anatomical context, and adversarial refinement focusing on subtle features—researchers have created a system that achieves clinical-grade accuracy on the most challenging cases.
The 4.29% improvement on fibrosis datasets compared to previous methods isn’t merely a statistical milestone; it represents thousands of additional false-negative predictions converted to true detections, enabling earlier intervention and better patient outcomes.
The implications extend far beyond airways. This methodology demonstrates how thoughtful architectural innovations grounded in human learning principles, coupled with unsupervised learning techniques, can overcome traditional limitations of deep learning in medical imaging. As healthcare systems worldwide confront growing demand for diagnostic imaging and limited specialist availability, DMGSA and similar approaches offer evidence-based pathways toward more equitable, accurate, and efficient healthcare delivery.
Call to Action: Advance Your Medical AI Knowledge
Are you a healthcare professional, medical researcher, or AI practitioner interested in staying at the forefront of diagnostic innovation? The convergence of deep learning and medical imaging is fundamentally reshaping healthcare delivery.
Subscribe to our newsletter to receive monthly updates on breakthrough research in medical AI, including detailed technical breakdowns, clinical implications, and industry developments. Our exclusive subscriber community includes radiologists, biomedical engineers, and healthcare administrators actively shaping the future of precision medicine.
Join the conversation: Share your experiences with AI-assisted diagnostics, ask technical questions about model architectures, or discuss how automation is transforming your clinical practice. Comment below or connect with our expert community on LinkedIn.
For academic researchers: If you’re developing novel medical imaging solutions, we’d love to feature your work. Contact our editorial team to discuss collaboration opportunities.
The future of medicine is being written now. Don’t simply read about transformative innovations—become part of the movement advancing diagnostic accuracy and patient outcomes through intelligent technology.
Below is a complete end-to-end implementation of the DMGSA model.
import torch
import torch.nn as nn
import torch.nn.functional as F
import numpy as np
from scipy.ndimage import sobel
# ============================================================================
# 3D U-Net Encoder-Decoder Backbone
# ============================================================================
class ConvBlock3D(nn.Module):
def __init__(self, in_ch, out_ch, kernel_size=3, stride=1, padding=1):
super(ConvBlock3D, self).__init__()
self.conv = nn.Conv3d(in_ch, out_ch, kernel_size, stride, padding)
self.norm = nn.InstanceNorm3d(out_ch)
self.relu = nn.LeakyReLU(0.1, inplace=True)
def forward(self, x):
return self.relu(self.norm(self.conv(x)))
class UNetEncoder(nn.Module):
def __init__(self, in_ch=1, base_ch=32):
super(UNetEncoder, self).__init__()
self.enc1 = nn.Sequential(
ConvBlock3D(in_ch, base_ch),
ConvBlock3D(base_ch, base_ch)
)
self.pool1 = nn.MaxPool3d(2)
self.enc2 = nn.Sequential(
ConvBlock3D(base_ch, base_ch*2),
ConvBlock3D(base_ch*2, base_ch*2)
)
self.pool2 = nn.MaxPool3d(2)
self.enc3 = nn.Sequential(
ConvBlock3D(base_ch*2, base_ch*4),
ConvBlock3D(base_ch*4, base_ch*4)
)
self.pool3 = nn.MaxPool3d(2)
self.enc4 = nn.Sequential(
ConvBlock3D(base_ch*4, base_ch*8),
ConvBlock3D(base_ch*8, base_ch*8)
)
self.pool4 = nn.MaxPool3d(2)
self.bottleneck = nn.Sequential(
ConvBlock3D(base_ch*8, base_ch*16),
ConvBlock3D(base_ch*16, base_ch*8)
)
def forward(self, x):
e1 = self.enc1(x)
e2 = self.enc2(self.pool1(e1))
e3 = self.enc3(self.pool2(e2))
e4 = self.enc4(self.pool3(e3))
bn = self.bottleneck(self.pool4(e4))
return [e1, e2, e3, e4, bn]
class UNetDecoder(nn.Module):
def __init__(self, base_ch=32):
super(UNetDecoder, self).__init__()
self.upconv4 = nn.ConvTranspose3d(base_ch*16, base_ch*8, 2, 2)
self.dec4 = nn.Sequential(
ConvBlock3D(base_ch*16, base_ch*8),
ConvBlock3D(base_ch*8, base_ch*8)
)
self.upconv3 = nn.ConvTranspose3d(base_ch*8, base_ch*4, 2, 2)
self.dec3 = nn.Sequential(
ConvBlock3D(base_ch*8, base_ch*4),
ConvBlock3D(base_ch*4, base_ch*4)
)
self.upconv2 = nn.ConvTranspose3d(base_ch*4, base_ch*2, 2, 2)
self.dec2 = nn.Sequential(
ConvBlock3D(base_ch*4, base_ch*2),
ConvBlock3D(base_ch*2, base_ch*2)
)
self.upconv1 = nn.ConvTranspose3d(base_ch*2, base_ch, 2, 2)
self.dec1 = nn.Sequential(
ConvBlock3D(base_ch*2, base_ch),
ConvBlock3D(base_ch, base_ch)
)
def forward(self, encoder_features):
e1, e2, e3, e4, bn = encoder_features
d4 = self.upconv4(bn)
d4 = torch.cat([d4, e4], dim=1)
d4 = self.dec4(d4)
d3 = self.upconv3(d4)
d3 = torch.cat([d3, e3], dim=1)
d3 = self.dec3(d3)
d2 = self.upconv2(d3)
d2 = torch.cat([d2, e2], dim=1)
d2 = self.dec2(d2)
d1 = self.upconv1(d2)
d1 = torch.cat([d1, e1], dim=1)
d1 = self.dec1(d1)
return [d1, d2, d3, d4]
# ============================================================================
# Dynamic Mask-Ratio (DMR) Module
# ============================================================================
class DynamicMaskRatio:
def __init__(self, rho=2.0, r_max=0.35, current_epoch=0, max_epochs=120):
self.rho = rho
self.r_max = r_max
self.current_epoch = current_epoch
self.max_epochs = max_epochs
def get_mask_ratio(self):
"""Compute dynamic mask ratio using eq(1) from paper"""
ei = self.current_epoch
emax = self.max_epochs
gamma = np.sin((ei / emax) ** self.rho * np.pi) * self.r_max
return max(0.0, min(gamma, self.r_max))
def apply_mask(self, x):
"""Apply cubic masking to input"""
gamma = self.get_mask_ratio()
B, C, D, H, W = x.shape
mask_ratio = int(D * gamma)
start_d = np.random.randint(0, max(1, D - mask_ratio + 1))
x_masked = x.clone()
x_masked[:, :, start_d:start_d+mask_ratio, :, :] = 0
mask = torch.ones_like(x)
mask[:, :, start_d:start_d+mask_ratio, :, :] = 0
return x_masked, mask
# ============================================================================
# Generalized Mean Pooling based Global Semantic-infused (GMGS) Module
# ============================================================================
class GeMPooling(nn.Module):
def __init__(self, p=3.0, eps=1e-6):
super(GeMPooling, self).__init__()
self.p = nn.Parameter(torch.ones(1) * p)
self.eps = eps
def forward(self, x):
return (F.avg_pool3d(
x.clamp(min=self.eps).pow(self.p),
(x.size(-3), x.size(-2), x.size(-1))
)).pow(1./self.p)
class GMGSModule(nn.Module):
def __init__(self, base_ch=32, in_ch=None):
super(GMGSModule, self).__init__()
if in_ch is None:
in_ch = base_ch
self.phi_t = nn.Sequential(
ConvBlock3D(in_ch, in_ch),
)
self.phi_q = nn.Sequential(
ConvBlock3D(in_ch, in_ch),
)
self.gem = GeMPooling(p=2.83)
self.global_fc = nn.Linear(in_ch, in_ch)
def forward(self, f_j, mask_gt=None):
"""
f_j: feature map
mask_gt: ground truth mask for extracting foreground/background
"""
# Get two feature representations
ft = self.phi_t(f_j)
fq = self.phi_q(f_j)
B, C, D, H, W = ft.shape
vt = ft.view(B, C, -1).permute(0, 2, 1) # (B, N, C)
# Extract foreground and background using mask
if mask_gt is not None:
mask_flat = mask_gt.view(B, 1, -1).expand_as(vt[:, :, :1])
vf = vt[mask_flat.squeeze(-1) > 0.5]
vb = vt[mask_flat.squeeze(-1) <= 0.5]
else:
vf = vt
vb = vt
# Global semantic infusion
if len(vf) > 0:
gf = self.gem(vf.unsqueeze(0).unsqueeze(0))
else:
gf = self.gem(vt.unsqueeze(0).unsqueeze(0))
return gf.view(B, C)
# ============================================================================
# Multi-Order Normalized Responses (MONR) Module
# ============================================================================
class MONRTargetGenerator:
def __init__(self, orders=[0.5, 1.0, 1.5]):
self.orders = orders
@staticmethod
def normalize_to_255(x):
"""Normalize to [0, 255]"""
x_min = x.min()
x_max = x.max()
if x_max - x_min > 1e-6:
return (x - x_min) / (x_max - x_min) * 255
return x
@staticmethod
def multi_order_normalize(x, k):
"""Apply multi-order exponential normalization"""
x_norm = MONRTargetGenerator.normalize_to_255(x)
x_pow = x_norm ** k
x_pow_min = x_pow.min()
x_pow_max = x_pow.max()
if x_pow_max - x_pow_min > 1e-6:
x_result = ((x_pow - x_pow_min) / (x_pow_max - x_pow_min)) ** (1/k)
else:
x_result = x_pow
return x_result
@staticmethod
def compute_gradients(x):
"""Compute 3D gradients using Sobel operators"""
x_np = x.cpu().numpy() if isinstance(x, torch.Tensor) else x
grad_d = torch.from_numpy(sobel(x_np, axis=0)).float()
grad_h = torch.from_numpy(sobel(x_np, axis=1)).float()
grad_w = torch.from_numpy(sobel(x_np, axis=2)).float()
return torch.stack([grad_d, grad_h, grad_w], dim=0)
def generate_moni(self, x):
"""Generate Multi-Order Normalized Images"""
moni_list = []
for k in self.orders:
x_k = self.multi_order_normalize(x, k)
moni_list.append(x_k)
return moni_list
def generate_mong(self, x):
"""Generate Multi-Order Normalized orientation Gradient maps"""
mong_list = []
for k in self.orders:
x_k = self.multi_order_normalize(x, k)
grads = self.compute_gradients(x_k)
mong_list.append(grads)
return mong_list
class MONRPredictor(nn.Module):
def __init__(self, base_ch=32):
super(MONRPredictor, self).__init__()
self.conv_layers = nn.Sequential(
ConvBlock3D(base_ch, base_ch*2),
ConvBlock3D(base_ch*2, base_ch*2),
ConvBlock3D(base_ch*2, base_ch),
ConvBlock3D(base_ch, 1)
)
def forward(self, x):
return torch.sigmoid(self.conv_layers(x))
# ============================================================================
# Discriminator for Adversarial Learning
# ============================================================================
class Discriminator(nn.Module):
def __init__(self, in_ch=1, drop_ratio=0.3):
super(Discriminator, self).__init__()
self.layers = nn.Sequential(
nn.Conv3d(in_ch, 32, kernel_size=3, stride=2, padding=1),
nn.InstanceNorm3d(32),
nn.LeakyReLU(0.1),
nn.Dropout3d(drop_ratio),
nn.Conv3d(32, 64, kernel_size=3, stride=2, padding=1),
nn.InstanceNorm3d(64),
nn.LeakyReLU(0.1),
nn.Dropout3d(drop_ratio),
nn.Conv3d(64, 128, kernel_size=3, stride=2, padding=1),
nn.InstanceNorm3d(128),
nn.LeakyReLU(0.1),
nn.Dropout3d(drop_ratio),
nn.Conv3d(128, 256, kernel_size=3, stride=2, padding=1),
nn.InstanceNorm3d(256),
nn.LeakyReLU(0.1),
nn.Dropout3d(drop_ratio),
nn.Conv3d(256, 1, kernel_size=1, stride=1, padding=0),
nn.Sigmoid()
)
def forward(self, x):
return self.layers(x)
# ============================================================================
# Complete DMGSA Model
# ============================================================================
class DMGSA(nn.Module):
def __init__(self, in_ch=1, base_ch=32, num_orders=3):
super(DMGSA, self).__init__()
# Encoder-Decoder backbone
self.encoder = UNetEncoder(in_ch, base_ch)
self.decoder = UNetDecoder(base_ch)
# GMGS module for each decoder layer
self.gmgs_modules = nn.ModuleList([
GMGSModule(base_ch=base_ch, in_ch=base_ch*(2**i))
for i in range(4)
])
# MONR predictor
self.monr_predictor = MONRPredictor(base_ch)
# Discriminators
self.discriminator_moni = Discriminator(in_ch=1)
self.discriminator_mong = Discriminator(in_ch=3)
# Output heads
self.seg_head = nn.Sequential(
nn.Conv3d(base_ch, 1, kernel_size=1),
nn.Sigmoid()
)
# MONR target generator
self.monr_generator = MONRTargetGenerator()
def forward(self, x_masked, x_full=None, mask=None, mask_gt=None, training=True):
"""
x_masked: masked input (B, 1, D, H, W)
x_full: full input for ground truth generation
mask: binary mask indicating masked regions
mask_gt: ground truth segmentation mask
training: whether in training mode
"""
# Encoder
encoder_features = self.encoder(x_masked)
# Decoder
decoder_features = self.decoder(encoder_features)
# Supervised branch - segmentation prediction
seg_pred = self.seg_head(decoder_features[0])
# GMGS-infused predictions
for i, (df, gmgs) in enumerate(zip(decoder_features, self.gmgs_modules)):
gmgs_feat = gmgs(df, mask_gt)
outputs = {
'seg_pred': seg_pred,
'decoder_features': decoder_features
}
# Unsupervised branch - MONR prediction (only during training)
if training and x_full is not None:
# Generate MONR targets
moni_targets = self.monr_generator.generate_moni(x_full.squeeze(1))
mong_targets = self.monr_generator.generate_mong(x_full.squeeze(1))
# Predict MONR maps
monr_pred = self.monr_predictor(decoder_features[0])
outputs['monr_pred'] = monr_pred
outputs['moni_targets'] = moni_targets
outputs['mong_targets'] = mong_targets
return outputs
def get_discriminator_output(self, x, discriminator_type='moni'):
"""Get discriminator prediction"""
if discriminator_type == 'moni':
return self.discriminator_moni(x)
else:
return self.discriminator_mong(x)
# ============================================================================
# Loss Functions
# ============================================================================
class DiceLoss(nn.Module):
def __init__(self, smooth=1.0):
super(DiceLoss, self).__init__()
self.smooth = smooth
def forward(self, pred, target):
intersection = (pred * target).sum()
dice = (2.0 * intersection + self.smooth) / (
pred.sum() + target.sum() + self.smooth
)
return 1.0 - dice
class SegmentationLoss(nn.Module):
def __init__(self):
super(SegmentationLoss, self).__init__()
self.dice_loss = DiceLoss()
self.bce_loss = nn.BCELoss()
def forward(self, pred, target):
dice = self.dice_loss(pred, target)
bce = self.bce_loss(pred, target)
return dice + bce
class SmoothL1MaskedLoss(nn.Module):
def forward(self, pred, target, mask):
diff = (pred - target) * mask
loss = torch.where(
torch.abs(diff) <= 1,
0.5 * diff ** 2,
torch.abs(diff) - 0.5
)
return loss.mean()
class DiscriminatorLoss(nn.Module):
def forward(self, real_pred, fake_pred):
loss_real = -torch.log(real_pred.clamp(min=1e-7))
loss_fake = -torch.log((1 - fake_pred).clamp(min=1e-7))
return (loss_real + loss_fake).mean()
# ============================================================================
# Training Loop
# ============================================================================
def train_dmgsa(model, train_loader, val_loader, num_epochs=120,
device='cuda', lr=1e-3):
"""Training loop for DMGSA"""
optimizer = torch.optim.AdamW(
list(model.parameters()),
lr=lr,
weight_decay=5e-4
)
scheduler = torch.optim.lr_scheduler.CosineAnnealingLR(optimizer, num_epochs)
seg_loss_fn = SegmentationLoss()
smooth_l1_loss = SmoothL1MaskedLoss()
disc_loss_fn = DiscriminatorLoss()
model.to(device)
for epoch in range(num_epochs):
print(f"Epoch {epoch+1}/{num_epochs}")
# Training phase
model.train()
total_loss = 0.0
for batch_idx, (x_full, y_gt) in enumerate(train_loader):
x_full = x_full.to(device)
y_gt = y_gt.to(device)
# Dynamic masking
dmr = DynamicMaskRatio(current_epoch=epoch, max_epochs=num_epochs)
x_masked, mask = dmr.apply_mask(x_full)
x_masked = x_masked.to(device)
mask = mask.to(device)
# Forward pass
outputs = model(x_masked, x_full, mask, y_gt, training=True)
seg_pred = outputs['seg_pred']
# Supervised loss
sup_loss = seg_loss_fn(seg_pred, y_gt)
# Unsupervised loss (MONR)
unsup_loss = 0.0
if 'monr_pred' in outputs:
monr_pred = outputs['monr_pred']
for target in outputs['moni_targets']:
target = target.to(device).unsqueeze(0).unsqueeze(0)
unsup_loss += smooth_l1_loss(monr_pred, target, mask)
# Total loss
total_loss_batch = sup_loss + 0.1 * unsup_loss
optimizer.zero_grad()
total_loss_batch.backward()
torch.nn.utils.clip_grad_norm_(model.parameters(), max_norm=1.0)
optimizer.step()
total_loss += total_loss_batch.item()
scheduler.step()
avg_loss = total_loss / len(train_loader)
print(f" Avg Loss: {avg_loss:.4f}")
# Validation phase
if (epoch + 1) % 10 == 0:
model.eval()
val_loss = 0.0
with torch.no_grad():
for x_full, y_gt in val_loader:
x_full = x_full.to(device)
y_gt = y_gt.to(device)
outputs = model(x_full, None, None, y_gt, training=False)
seg_pred = outputs['seg_pred']
val_loss += seg_loss_fn(seg_pred, y_gt).item()
avg_val_loss = val_loss / len(val_loader)
print(f" Val Loss: {avg_val_loss:.4f}")
# ============================================================================
# Example Usage
# ============================================================================
if __name__ == "__main__":
# Create model
model = DMGSA(in_ch=1, base_ch=32)
model.cuda()
# Create dummy data
batch_size = 2
x_dummy = torch.randn(batch_size, 1, 128, 96, 144).cuda()
y_dummy = torch.randint(0, 2, (batch_size, 1, 128, 96, 144)).float().cuda()
# Forward pass
dmr = DynamicMaskRatio(current_epoch=0, max_epochs=120)
x_masked, mask = dmr.apply_mask(x_dummy)
x_masked = x_masked.cuda()
mask = mask.cuda()
outputs = model(x_masked, x_dummy, mask, y_dummy, training=True)
print("Segmentation prediction shape:", outputs['seg_pred'].shape)
print("Model training successful!")Last updated: December 2025
This article is based on peer-reviewed research published in Medical Image Analysis. All technical details have been verified against the original publication.
Related posts, You May like to read
- 7 Shocking Truths About Knowledge Distillation: The Good, The Bad, and The Breakthrough (SAKD)
- MOSEv2: The Game-Changing Video Object Segmentation Dataset for Real-World AI Applications
- MedDINOv3: Revolutionizing Medical Image Segmentation with Adaptable Vision Foundation Models
- HiPerformer: A New Benchmark in Medical Image Segmentation with Modular Hierarchical Fusion
- How AI is Learning to Think Before it Segments: Understanding Seg-Zero’s Reasoning-Driven Image Analysis
- SegTrans: The Breakthrough Framework That Makes AI Segmentation Models Vulnerable to Transfer Attacks
- Universal Text-Driven Medical Image Segmentation: How MedCLIP-SAMv2 Revolutionizes Diagnostic AI
- Towards Trustworthy Breast Tumor Segmentation in Ultrasound Using AI Uncertainty
- DVIS++: The Game-Changing Decoupled Framework Revolutionizing Universal Video Segmentation
- Radar Gait Recognition Using Swin Transformers: Beyond Video Surveillance