Introduction
Cancer remains one of the leading causes of mortality worldwide, yet advances in personalized medicine and artificial intelligence are fundamentally transforming how physicians predict patient survival and recommend treatment strategies. Traditional prognostic approaches rely on limited clinical variables and single-source data, often missing the complex biological heterogeneity that characterizes modern cancer. Recent breakthroughs in computational pathology and multimodal machine learning are changing this landscape.
Researchers have developed M³Surv, a sophisticated artificial intelligence framework that integrates histopathological images with comprehensive genomic, transcriptomic, and proteomic data to deliver unprecedented accuracy in cancer survival prediction. What makes this innovation particularly significant is its ability to function effectively even when critical data is missing—a real-world challenge that has long plagued multimodal AI systems in clinical settings.
This comprehensive article explores the groundbreaking methodology behind M³Surv, its clinical implications, and why this technology represents a paradigm shift in precision oncology.
Understanding the Challenge: Why Traditional Survival Prediction Falls Short
The Complexity of Cancer Heterogeneity
Cancer is inherently heterogeneous. Two patients diagnosed with the same cancer type can exhibit vastly different molecular profiles, treatment responses, and survival outcomes. Traditional survival prediction models often rely on basic clinical factors—tumor grade, size, and stage—which provide only a superficial understanding of disease biology.
Modern cancer research has revealed that patient prognosis depends on intricate interactions between:
- Tumor morphology (cellular architecture visible in tissue slides)
- Gene expression patterns (which genes are active)
- Genomic mutations (DNA-level alterations)
- Protein abundance (functional molecular components)
The fundamental problem: No single data source captures all this information. Histopathology images reveal cellular organization but lack molecular details. Gene sequencing provides molecular insights but misses morphological context. Survival prediction requires synthesizing all these dimensions simultaneously.
The Missing Data Problem in Real-World Clinics
Hospital laboratories face genuine resource constraints. Preparing Fresh-Frozen (FF) tissue samples takes 15 minutes, while Formalin-Fixed Paraffin-Embedded (FFPE) processing requires approximately 36 hours. Genetic sequencing can be expensive, transcriptomic analysis demanding, and proteomic profiling resource-intensive. In practice, not every patient has complete multimodal data available.
Existing multimodal AI systems break down when data is incomplete. They rely on inter-modality correlations that disappear when an entire data type is missing. This creates a critical clinical gap: the most sophisticated predictive tools become unusable precisely when circumstances prevent complete data collection.
M³Surv: A Breakthrough Framework for Robust Multimodal Prediction
The Three Pillars of M³Surv
M³Surv addresses these challenges through three innovative components:
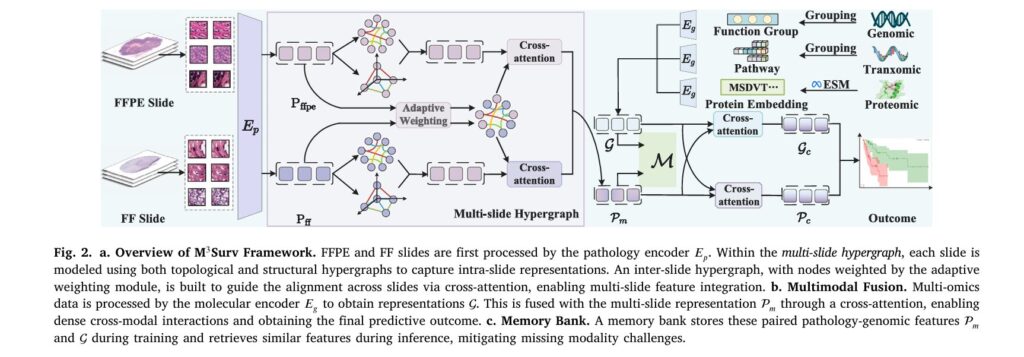
1. Multi-Slide Hypergraph Learning
Rather than treating FF and FFPE slides as separate entities, M³Surv employs sophisticated hypergraph neural networks to model them as complementary views of the same patient’s tissue.
Hypergraphs extend traditional graph theory by allowing a single edge (called a hyperedge) to connect multiple nodes simultaneously. This is more realistic for biological tissue, where clusters of similar cells interact together rather than in simple pairwise relationships.
The framework constructs two types of hyperedges within each slide:
- Topological hyperedges: Connect adjacent patches based on spatial proximity, capturing local tissue architecture
- Structural hyperedges: Link patches with similar morphological features, identifying semantically similar regions regardless of location
These intra-slide representations are then connected through inter-slide hyperedges that align semantically similar regions across FF and FFPE samples from the same patient. This ensures the model recognizes that the same biological structures appear in both slide types, despite staining and preparation differences.
Mathematical representation:
For a given patch pi, topological neighbors are identified as:
$$\mathcal{N}T(p_i) = {p_j \mid |\zeta{p_j} – \zeta_{p_i}|_2 \leq \delta}$$where ζpi and ζpj denote spatial coordinates and δ is a distance threshold.
Similarly, structural neighbors based on feature similarity are:
$$\mathcal{N}_F(p_i) = {p_j \mid \text{sim}(p_i, p_j) \geq \gamma}$$
2. Comprehensive Multi-Omics Integration
M³Surv moves beyond single-modality genomics analysis by integrating three complementary molecular layers:
- Genomics: Copy number variations and RNA-seq data grouped into six functional categories (Tumor Suppression, Oncogenesis, Protein Kinases, Cellular Differentiation, Transcription, and Cytokines/Growth)
- Transcriptomics: 331 biological pathways from Reactome and MSigDB databases, capturing dynamic gene expression across known cellular processes
- Proteomics: Top 100 proteins selected by expression level, with sequence embeddings generated through evolutionary-scale modeling
This comprehensive approach captures biology at multiple regulatory layers. Genomic alterations represent permanent mutations; transcriptomic changes reflect current gene activity; proteomic profiles represent the actual functional molecules driving cell behavior. Together, they paint a complete molecular picture unavailable through any single modality.
3. Prototype-Based Memory Bank for Missing Modality Imputation
Perhaps the most clinically valuable innovation is the memory bank—a learned repository of representative pathology-omics feature patterns discovered during training.
During training, the system clusters paired pathology-genomic features using k-means clustering, generating modality-specific prototypes that represent distinct patient subgroups. These prototypes are stored alongside their features in the memory bank with momentum-based updates that refine them across epochs.
The inference magic: When a modality is missing during prediction, the model computes a prototype from available data and queries the memory bank for the most similar stored prototype. It then retrieves the missing modality’s corresponding features from the most similar patients in the training cohort:
$$\widehat{M}m = \sum{j=1}^{\mu} \frac{\exp(\text{sim}(\mathbf{C}_a, \mathbf{C}a^{(j)}))}{\sum{h=1}^{\mu} \exp(\text{sim}(\mathbf{C}_a, \mathbf{C}_a^{(h)}))} M_m^{(j)}$$This similarity-weighted imputation is more robust than trying to predict missing data from a single patient’s partial information, because it leverages patterns from the entire training population.
Clinical Evidence: M³Surv Outperforms State-of-the-Art
Extensive experiments across multiple cancer types demonstrate M³Surv’s superior predictive power:
Performance on Five TCGA Datasets
Evaluation across five large The Cancer Genome Atlas (TCGA) cohorts shows consistent improvements:
| Cancer Type | Sample Size | M³Surv C-Index | Previous SOTA | Improvement |
|---|---|---|---|---|
| Bladder Carcinoma (BLCA) | 384 | 67.4% | 65.1% | +2.3% |
| Breast Carcinoma (BRCA) | 968 | 73.2% | 72.7% | +0.5% |
| Colorectal Adenocarcinoma (CRAD) | 298 | 78.8% | 78.6% | +0.2% |
| Head/Neck Cancer (HNSC) | 392 | 66.4% | 65.6% | +0.8% |
| Gastric Adenocarcinoma (STAD) | 317 | 67.7% | 66.8% | +0.9% |
| Average | 2359 | 70.7% | 68.5% | +2.2% |
The Concordance Index (C-Index) measures agreement between predicted risk scores and observed survival, ranging from 0.5 (random prediction) to 1.0 (perfect prediction). A 2.2% average improvement represents a clinically meaningful enhancement in prognostic accuracy.
Robustness Across Missing Data Scenarios
The memory bank’s effectiveness becomes apparent when evaluating performance under realistic clinical constraints:
With missing pathology data:
- 30% missing: 65.8% C-Index
- 50% missing: 67.3% C-Index
- 70% missing: 66.5% C-Index
- 100% missing: 66.9% C-Index
With missing multi-omics data:
- 30% missing: 68.0% C-Index
- 50% missing: 67.4% C-Index
- 70% missing: 68.5% C-Index
- 100% missing: 66.6% C-Index
Remarkably, performance at 100% missing data (where only one modality exists) often exceeded intermediate missing scenarios. This counterintuitive finding reveals that the memory bank works most effectively with complete information from a single modality, avoiding confusion from contradictory partial information.
Validation on In-House Data
Testing on an Anhui Provincial Hospital liver cancer cohort (APH-LC, n=302) corroborated findings from public datasets, confirming that M³Surv’s benefits generalize beyond academic research settings to real clinical populations.
Key Technical Innovations Driving Performance
Divide-and-Conquer Multi-Slide Processing
Rather than forcing FF and FFPE slides into a single representation, M³Surv employs a divide-and-conquer strategy:
- Aggregation: Each slide type is processed independently through hypergraph convolution layers, capturing slide-specific patterns
- Alignment: Cross-attention mechanisms refine FF and FFPE features using inter-slide features as shared guidance, ensuring biological consistency
- Integration: Refined representations are concatenated into a unified patient-level pathology representation
This approach respects the unique characteristics of each slide type—FFPE slides preserve morphology better; FF slides preserve molecular information better—while exploiting their complementarity at the patient level.
Adaptive Weighting for Dynamic Slide Contribution
An often-overlooked challenge: slides vary in diagnostic relevance. A single hypergene region might carry more prognostic weight than surrounding normal tissue. M³Surv addresses this through adaptive weighting:
$$w_{ff}, w_{ffpe} = \frac{\exp(W_{ff}P_{ff}), \exp(W_{ffpe}P_{ffpe})}{\exp(W_{ff}P_{ff}) + \exp(W_{ffpe}P_{ffpe})}$$Trainable weight matrices automatically learn which slides matter most for each patient, capturing disease heterogeneity at the individual level.
Mutual Cross-Attention for Bidirectional Fusion
Multimodal fusion typically privileges one modality as “primary.” M³Surv instead employs mutual cross-attention, where pathology and omics modalities enhance each other bidirectionally:
- Pathology enhanced by omics:
- Omics enhanced by pathology:
Both enhanced representations are concatenated and processed together, ensuring neither modality dominates the final prediction.
Interpretability: Understanding Why M³Surv Works
Visualization of Pathological Attention
Heatmap visualizations reveal that M³Surv’s hypergraph learning identifies broader, more coherent activation regions compared to baseline approaches. Rather than scattered high-attention patches, the model recognizes meaningful histological patterns spanning multiple tissue regions.
Biological Pathway Interpretation
For breast cancer patients, the framework identified Complement System and Iron Transport pathways as highly influential—patterns directly reflecting BRCA’s complex immune microenvironment and nutrient-demanding proliferation. Conversely, suppressed Apoptotic Cleavage pathways indicated loss of cell death mechanisms, a known hallmark of malignancy.
This interpretability is crucial for clinical adoption. Physicians can understand not just that M³Surv predicts poor survival, but why, facilitating confident treatment decisions.
Kaplan-Meier Stratification Power
Across all five TCGA datasets, M³Surv achieved log-rank test p-values < 0.05, confirming it genuinely separates high-risk from low-risk populations—not merely achieving statistical discrimination on academic metrics.
Clinical Implementation and Future Directions
Computational Efficiency
A common concern with advanced AI: computational cost. M³Surv adds only 4.3% overhead to training time and 2.2% to inference time through its memory bank, making deployment in busy hospital workflows feasible.
Path to Clinical Deployment
For M³Surv to transform oncology practice, several steps remain:
- Prospective clinical trials validating that predicted risk scores actually drive better treatment decisions and improved outcomes
- Integration with hospital information systems, making predictions available alongside standard pathology reports
- Interpretability enhancements, developing tools that explain predictions in language clinicians understand
- Regulatory approval, navigating FDA or equivalent pathways for AI-based clinical decision support
Promising Extensions
Researchers suggest several promising directions:
- Multimodal expansion: Incorporating radiology images and clinical notes alongside pathology and genomics
- Longitudinal monitoring: Updating survival predictions dynamically as new patient data become available
- Temporal dynamics: Modeling how molecular features evolve throughout treatment, enabling mid-course prognosis adjustment
What This Means for Patients and Clinicians
For patients, M³Surv offers more accurate prognosis, enabling better-informed discussions about treatment intensity, clinical trial eligibility, and life planning decisions.
For oncologists, it provides a principled second opinion integrating vast amounts of complex data that no human could synthesize manually. The ability to function with incomplete data is particularly valuable—clinicians need not delay treatment while awaiting expensive molecular tests.
For researchers, it demonstrates that respecting biological complexity—through multi-slide integration and comprehensive omics—yields real performance gains while remaining computationally practical.
The Bottom Line: A Paradigm Shift in Precision Oncology
M³Surv represents a genuine advance in how artificial intelligence can enhance clinical cancer care. By thoughtfully integrating multiple data modalities, intelligently handling real-world incompleteness, and maintaining interpretability, it achieves what previous approaches could not: a robust, practical, accurate survival prediction system suitable for heterogeneous patient populations and imperfect clinical environments.
The 2.2% average improvement in predictive accuracy might seem modest in isolation. But in oncology—where each percentage point improvement in risk stratification translates to more targeted treatment, fewer unnecessary toxic therapies, and better patient outcomes—this represents meaningful clinical progress.
As this technology matures through clinical validation and integration into hospital workflows, it will likely join the arsenal of tools that drive increasingly personalized, data-driven cancer care.
Ready to Understand More About AI in Healthcare?
The intersection of artificial intelligence and clinical medicine is evolving rapidly. If you work in oncology, computational pathology, or healthcare AI, understanding frameworks like M³Surv is essential for staying current with the field’s trajectory.
Share your thoughts: Are you involved in developing or implementing AI for cancer prognostication? Have you encountered challenges with multimodal data integration in clinical settings? Leave a comment below or reach out to discuss how these technologies might apply to your specific clinical or research context.
Stay informed: Subscribe to receive updates on emerging developments in precision oncology and healthcare AI—the field advances faster than ever before.
If you want read the full paper, then click the link here.
Below is a complete, end-to-end implementation of the M³Surv framework with all major components.
"""
M³Surv: Memory-Augmented Multimodal Survival Prediction Framework
Complete end-to-end implementation with multi-slide hypergraph learning,
multi-omics integration, and prototype-based memory bank for handling missing modalities.
"""
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.optim import Adam
from torch_geometric.nn import GCNConv, HypergraphConv
import numpy as np
from sklearn.cluster import KMeans
from sklearn.preprocessing import StandardScaler
from typing import Dict, List, Tuple, Optional
import warnings
warnings.filterwarnings('ignore')
# ============================================================================
# FEATURE EXTRACTION ENCODERS
# ============================================================================
class PathologyEncoder(nn.Module):
"""
Encodes pathology features from WSI patches using ResNet50 backbone.
Extracts d-dimensional features from each patch at 20x magnification.
"""
def __init__(self, feature_dim: int = 768, pretrained: bool = True):
super(PathologyEncoder, self).__init__()
import torchvision.models as models
resnet = models.resnet50(pretrained=pretrained)
self.backbone = nn.Sequential(*list(resnet.children())[:-1])
self.feature_dim = feature_dim
self.projection = nn.Linear(2048, feature_dim)
def forward(self, patch_features: torch.Tensor) -> torch.Tensor:
"""
Args:
patch_features: (N_patches, 3, 256, 256) image tensor
Returns:
embeddings: (N_patches, feature_dim) feature embeddings
"""
x = self.backbone(patch_features) # (N_patches, 2048, 1, 1)
x = x.squeeze(-1).squeeze(-1) # (N_patches, 2048)
embeddings = self.projection(x) # (N_patches, feature_dim)
return embeddings
class GenomicsEncoder(nn.Module):
"""
Encodes genomic data into functional categories using Spiking Neural Networks.
Maps 6 functional gene groups to feature space.
"""
def __init__(self, feature_dim: int = 768):
super(GenomicsEncoder, self).__init__()
self.feature_dim = feature_dim
# 6 functional gene categories
self.genomics_proj = nn.Linear(6, feature_dim)
self.norm = nn.BatchNorm1d(feature_dim)
def forward(self, genomics: torch.Tensor) -> torch.Tensor:
"""
Args:
genomics: (batch_size, 6) functional gene features
Returns:
embeddings: (batch_size, feature_dim)
"""
x = self.genomics_proj(genomics)
x = self.norm(x)
return x
class TranscriptomicsEncoder(nn.Module):
"""
Encodes transcriptomic pathway data from Reactome and MSigDB.
Maps 331 biological pathways to feature space.
"""
def __init__(self, num_pathways: int = 331, feature_dim: int = 768):
super(TranscriptomicsEncoder, self).__init__()
self.feature_dim = feature_dim
self.pathway_embedding = nn.Linear(num_pathways, feature_dim)
self.norm = nn.BatchNorm1d(feature_dim)
def forward(self, transcriptomics: torch.Tensor) -> torch.Tensor:
"""
Args:
transcriptomics: (batch_size, 331) pathway expression
Returns:
embeddings: (batch_size, feature_dim)
"""
x = self.pathway_embedding(transcriptomics)
x = self.norm(x)
return x
class ProteomicsEncoder(nn.Module):
"""
Encodes protein expression and sequence information.
Uses protein sequence embeddings (ESM) combined with expression levels.
"""
def __init__(self, num_proteins: int = 100, feature_dim: int = 768):
super(ProteomicsEncoder, self).__init__()
self.feature_dim = feature_dim
self.protein_embedding = nn.Linear(num_proteins + 1280, feature_dim) # 1280 = ESM embed dim
self.norm = nn.BatchNorm1d(feature_dim)
def forward(self, protein_expr: torch.Tensor,
protein_seq_embed: torch.Tensor) -> torch.Tensor:
"""
Args:
protein_expr: (batch_size, 100) protein expression levels
protein_seq_embed: (batch_size, 100, 1280) sequence embeddings
Returns:
embeddings: (batch_size, feature_dim)
"""
# Average sequence embeddings across proteins
seq_avg = protein_seq_embed.mean(dim=1) # (batch_size, 1280)
combined = torch.cat([protein_expr, seq_avg], dim=1) # (batch_size, 1380)
x = self.protein_embedding(combined)
x = self.norm(x)
return x
# ============================================================================
# HYPERGRAPH LEARNING MODULES
# ============================================================================
class HypergraphConstruction(nn.Module):
"""
Constructs intra-slide hypergraphs with topological and structural hyperedges.
Topological: spatial proximity (Euclidean distance)
Structural: morphological similarity (cosine distance)
"""
def __init__(self, spatial_threshold: float = 100.0,
similarity_threshold: float = 0.7, k_neighbors: int = 9):
super(HypergraphConstruction, self).__init__()
self.spatial_threshold = spatial_threshold
self.similarity_threshold = similarity_threshold
self.k_neighbors = k_neighbors
def forward(self, patches: torch.Tensor,
coordinates: torch.Tensor) -> Tuple[torch.Tensor, torch.Tensor]:
"""
Constructs hyperedge indices for topological and structural connections.
Args:
patches: (N_patches, feature_dim)
coordinates: (N_patches, 2) spatial coordinates
Returns:
topo_edges: topological hyperedge indices
struct_edges: structural hyperedge indices
"""
N = patches.shape[0]
device = patches.device
# Topological hyperedges (spatial proximity)
distances = torch.cdist(coordinates, coordinates) # (N, N)
topo_mask = distances <= self.spatial_threshold
topo_edges = torch.nonzero(topo_mask, as_tuple=False).t() # (2, num_edges)
# Structural hyperedges (feature similarity)
# Normalize patches for cosine similarity
patches_norm = F.normalize(patches, p=2, dim=1)
similarity = torch.mm(patches_norm, patches_norm.t()) # (N, N)
struct_mask = similarity >= self.similarity_threshold
struct_edges = torch.nonzero(struct_mask, as_tuple=False).t()
return topo_edges.to(device), struct_edges.to(device)
class IntraSlideHypergraphConv(nn.Module):
"""
Hypergraph convolution layer for intra-slide feature aggregation.
Captures higher-order cellular structures through hyperedges.
"""
def __init__(self, in_channels: int, out_channels: int, num_layers: int = 3):
super(IntraSlideHypergraphConv, self).__init__()
self.layers = nn.ModuleList([
HypergraphConv(in_channels if i == 0 else out_channels, out_channels)
for i in range(num_layers)
])
self.norm = nn.BatchNorm1d(out_channels)
self.relu = nn.ReLU()
def forward(self, x: torch.Tensor, hyperedge_index: torch.Tensor) -> torch.Tensor:
"""
Args:
x: (N_patches, in_channels) node features
hyperedge_index: (2, num_edges) hyperedge indices
Returns:
aggregated: (N_patches, out_channels)
"""
for layer in self.layers:
x = layer(x, hyperedge_index)
x = self.norm(x)
x = self.relu(x)
return x
class InterSlideHypergraph(nn.Module):
"""
Models relationships between FF and FFPE slides using inter-slide hyperedges.
Aligns semantically similar patches across slide types.
"""
def __init__(self, feature_dim: int = 768, similarity_threshold: float = 0.8):
super(InterSlideHypergraph, self).__init__()
self.similarity_threshold = similarity_threshold
self.feature_dim = feature_dim
def forward(self, ff_patches: torch.Tensor,
ffpe_patches: torch.Tensor) -> Tuple[torch.Tensor, torch.Tensor]:
"""
Identifies inter-slide correspondences based on feature similarity.
Args:
ff_patches: (N_ff, feature_dim)
ffpe_patches: (N_ffpe, feature_dim)
Returns:
inter_edges: hyperedge indices
similarity_scores: alignment confidence scores
"""
# Normalize for cosine similarity
ff_norm = F.normalize(ff_patches, p=2, dim=1)
ffpe_norm = F.normalize(ffpe_patches, p=2, dim=1)
# Cross-slide similarity
cross_similarity = torch.mm(ff_norm, ffpe_norm.t()) # (N_ff, N_ffpe)
# Find high-confidence matches
mask = cross_similarity >= self.similarity_threshold
inter_edges = torch.nonzero(mask, as_tuple=False).t()
sim_scores = cross_similarity[mask]
return inter_edges, sim_scores
# ============================================================================
# MULTIMODAL FUSION
# ============================================================================
class CrossAttentionFusion(nn.Module):
"""
Bidirectional cross-attention fusion between pathology and omics modalities.
Enables rich inter-modality interactions.
"""
def __init__(self, feature_dim: int = 768, num_heads: int = 8):
super(CrossAttentionFusion, self).__init__()
self.feature_dim = feature_dim
self.num_heads = num_heads
# Query, Key, Value projections for pathology -> omics
self.W_q_p = nn.Linear(feature_dim, feature_dim)
self.W_k_g = nn.Linear(feature_dim, feature_dim)
self.W_v_g = nn.Linear(feature_dim, feature_dim)
# Query, Key, Value projections for omics -> pathology
self.W_q_g = nn.Linear(feature_dim, feature_dim)
self.W_k_p = nn.Linear(feature_dim, feature_dim)
self.W_v_p = nn.Linear(feature_dim, feature_dim)
self.scale = np.sqrt(feature_dim)
self.softmax = nn.Softmax(dim=-1)
def forward(self, pathology: torch.Tensor,
omics: torch.Tensor) -> Tuple[torch.Tensor, torch.Tensor]:
"""
Args:
pathology: (batch_size, num_patches, feature_dim) or (batch_size, feature_dim)
omics: (batch_size, num_omic_features, feature_dim) or (batch_size, feature_dim)
Returns:
pathology_enhanced: pathology attended to by omics
omics_enhanced: omics attended to by pathology
"""
# Handle 2D inputs (flattened features)
if pathology.dim() == 2:
pathology = pathology.unsqueeze(1)
if omics.dim() == 2:
omics = omics.unsqueeze(1)
# Pathology attended to by omics (multi-head simulation)
Q_p = self.W_q_p(pathology) # (batch, seq_p, feature_dim)
K_g = self.W_k_g(omics) # (batch, seq_g, feature_dim)
V_g = self.W_v_g(omics) # (batch, seq_g, feature_dim)
scores_pg = torch.matmul(Q_p, K_g.transpose(-2, -1)) / self.scale
attn_pg = self.softmax(scores_pg)
pathology_enhanced = torch.matmul(attn_pg, V_g) # (batch, seq_p, feature_dim)
# Omics attended to by pathology
Q_g = self.W_q_g(omics)
K_p = self.W_k_p(pathology)
V_p = self.W_v_p(pathology)
scores_gp = torch.matmul(Q_g, K_p.transpose(-2, -1)) / self.scale
attn_gp = self.softmax(scores_gp)
omics_enhanced = torch.matmul(attn_gp, V_p) # (batch, seq_g, feature_dim)
# Flatten back to (batch, feature_dim)
pathology_enhanced = pathology_enhanced.mean(dim=1)
omics_enhanced = omics_enhanced.mean(dim=1)
return pathology_enhanced, omics_enhanced
# ============================================================================
# PROTOTYPE-BASED MEMORY BANK
# ============================================================================
class PrototypeMemoryBank(nn.Module):
"""
Memory bank storing representative pathology-omics feature prototypes.
Enables robust imputation for missing modalities during inference.
"""
def __init__(self, num_prototypes: int = 10, feature_dim: int = 768,
momentum: float = 0.75):
super(PrototypeMemoryBank, self).__init__()
self.num_prototypes = num_prototypes
self.feature_dim = feature_dim
self.momentum = momentum
# Initialize memory bank
self.register_buffer('proto_pathology',
torch.randn(num_prototypes, feature_dim) / np.sqrt(feature_dim))
self.register_buffer('proto_omics',
torch.randn(num_prototypes, feature_dim) / np.sqrt(feature_dim))
self.pathology_features_buffer = []
self.omics_features_buffer = []
self.cluster_assignments = []
def update_prototypes(self, pathology_feats: torch.Tensor,
omics_feats: torch.Tensor):
"""
Updates prototypes via momentum-based update using k-means clustering.
Args:
pathology_feats: (batch_size, feature_dim)
omics_feats: (batch_size, feature_dim)
"""
device = pathology_feats.device
# Store features
self.pathology_features_buffer.append(pathology_feats.detach().cpu().numpy())
self.omics_features_buffer.append(omics_feats.detach().cpu().numpy())
# Perform k-means clustering periodically (every N batches)
if len(self.pathology_features_buffer) >= 5:
# Concatenate all stored features
all_path = np.concatenate(self.pathology_features_buffer, axis=0)
all_omics = np.concatenate(self.omics_features_buffer, axis=0)
# Cluster pathology features
kmeans_p = KMeans(n_clusters=self.num_prototypes, random_state=42, n_init=10)
kmeans_p.fit(all_path)
proto_path_new = torch.tensor(kmeans_p.cluster_centers_,
dtype=torch.float32, device=device)
# Cluster omics features
kmeans_g = KMeans(n_clusters=self.num_prototypes, random_state=42, n_init=10)
kmeans_g.fit(all_omics)
proto_omics_new = torch.tensor(kmeans_g.cluster_centers_,
dtype=torch.float32, device=device)
# Momentum update
self.proto_pathology.data = (self.momentum * self.proto_pathology.data +
(1 - self.momentum) * proto_path_new)
self.proto_omics.data = (self.momentum * self.proto_omics.data +
(1 - self.momentum) * proto_omics_new)
# Clear buffers
self.pathology_features_buffer = []
self.omics_features_buffer = []
def retrieve_missing_modality(self, available_feats: torch.Tensor,
missing_type: str = 'omics',
top_k: int = 1) -> torch.Tensor:
"""
Retrieves missing modality features from memory bank using available modality.
Args:
available_feats: (batch_size, feature_dim) available modality features
missing_type: 'omics' or 'pathology'
top_k: number of prototypes to retrieve and average
Returns:
imputed_feats: (batch_size, feature_dim) imputed features
"""
available_feats_norm = F.normalize(available_feats, p=2, dim=1)
if missing_type == 'omics':
# Available: pathology, retrieve: omics
proto_available = F.normalize(self.proto_pathology, p=2, dim=1)
proto_missing = self.proto_omics
else:
# Available: omics, retrieve: pathology
proto_available = F.normalize(self.proto_omics, p=2, dim=1)
proto_missing = self.proto_pathology
# Similarity-weighted retrieval
similarity = torch.mm(available_feats_norm, proto_available.t()) # (batch, num_proto)
top_sim, top_indices = torch.topk(similarity, k=min(top_k, self.num_prototypes), dim=1)
# Softmax weights for weighted combination
weights = F.softmax(top_sim, dim=1) # (batch, top_k)
# Weighted sum of retrieved prototypes
imputed = torch.zeros_like(available_feats)
for b in range(available_feats.shape[0]):
imputed[b] = (weights[b].unsqueeze(1) *
proto_missing[top_indices[b]]).sum(dim=0)
return imputed
# ============================================================================
# SURVIVAL PREDICTION HEAD
# ============================================================================
class SurvivalHead(nn.Module):
"""
Predicts hazard and survival functions using negative log-likelihood loss.
Supports discrete-time survival analysis.
"""
def __init__(self, feature_dim: int = 768, num_time_bins: int = 50,
hidden_dim: int = 256):
super(SurvivalHead, self).__init__()
self.feature_dim = feature_dim
self.num_time_bins = num_time_bins
# Prediction network
self.fc1 = nn.Linear(feature_dim, hidden_dim)
self.dropout = nn.Dropout(0.2)
self.fc2 = nn.Linear(hidden_dim, hidden_dim)
# Hazard and survival outputs
self.hazard_head = nn.Linear(hidden_dim, num_time_bins)
self.survival_head = nn.Linear(hidden_dim, num_time_bins)
self.relu = nn.ReLU()
def forward(self, features: torch.Tensor) -> Tuple[torch.Tensor, torch.Tensor]:
"""
Args:
features: (batch_size, feature_dim) fused features
Returns:
hazard: (batch_size, num_time_bins)
survival: (batch_size, num_time_bins)
"""
x = self.fc1(features)
x = self.relu(x)
x = self.dropout(x)
x = self.fc2(x)
x = self.relu(x)
hazard = self.hazard_head(x)
survival = F.sigmoid(self.survival_head(x)) # Constrain to [0, 1]
return hazard, survival
# ============================================================================
# MAIN M³SURV MODEL
# ============================================================================
class M3Surv(nn.Module):
"""
Complete M³Surv framework integrating:
- Multi-slide hypergraph learning (FF + FFPE)
- Multi-omics fusion (genomics + transcriptomics + proteomics)
- Prototype-based memory bank for missing modality imputation
"""
def __init__(self, feature_dim: int = 768, num_time_bins: int = 50,
num_prototypes: int = 10, momentum: float = 0.75,
spatial_threshold: float = 100.0,
similarity_threshold: float = 0.7):
super(M3Surv, self).__init__()
self.feature_dim = feature_dim
self.num_time_bins = num_time_bins
# Feature encoders
self.pathology_encoder = PathologyEncoder(feature_dim)
self.genomics_encoder = GenomicsEncoder(feature_dim)
self.transcriptomics_encoder = TranscriptomicsEncoder(feature_dim=feature_dim)
self.proteomics_encoder = ProteomicsEncoder(feature_dim=feature_dim)
# Hypergraph modules
self.hypergraph_construction = HypergraphConstruction(
spatial_threshold=spatial_threshold,
similarity_threshold=similarity_threshold
)
self.intra_slide_hypergraph = IntraSlideHypergraphConv(
feature_dim, feature_dim, num_layers=3
)
self.inter_slide_hypergraph = InterSlideHypergraph(
feature_dim=feature_dim,
similarity_threshold=0.8
)
# Adaptive slide weighting
self.slide_weight_ff = nn.Parameter(torch.ones(feature_dim, 1) * 0.5)
self.slide_weight_ffpe = nn.Parameter(torch.ones(feature_dim, 1) * 0.5)
# Cross-attention fusion
self.cross_attention = CrossAttentionFusion(feature_dim, num_heads=8)
# Memory bank for missing modality handling
self.memory_bank = PrototypeMemoryBank(
num_prototypes=num_prototypes,
feature_dim=feature_dim,
momentum=momentum
)
# Survival prediction head
self.survival_head = SurvivalHead(feature_dim * 2, num_time_bins)
def forward(self, ff_patches: torch.Tensor, ffpe_patches: torch.Tensor,
ff_coords: torch.Tensor, ffpe_coords: torch.Tensor,
genomics: torch.Tensor, transcriptomics: torch.Tensor,
proteomics: torch.Tensor, protein_seq_embed: torch.Tensor,
missing_modality: Optional[str] = None) -> Tuple[torch.Tensor, torch.Tensor]:
"""
Complete forward pass through M³Surv.
Args:
ff_patches: (batch_size, N_ff_patches, 3, 256, 256) or (batch_size, feat_dim)
ffpe_patches: (batch_size, N_ffpe_patches, 3, 256, 256) or (batch_size, feat_dim)
ff_coords: (batch_size, N_ff_patches, 2)
ffpe_coords: (batch_size, N_ffpe_patches, 2)
genomics: (batch_size, 6)
transcriptomics: (batch_size, 331)
proteomics: (batch_size, 100)
protein_seq_embed: (batch_size, 100, 1280)
missing_modality: None, 'pathology', or 'omics'
Returns:
hazard: (batch_size, num_time_bins)
survival: (batch_size, num_time_bins)
"""
batch_size = genomics.shape[0]
device = genomics.device
# ===== PATHOLOGY FEATURE EXTRACTION =====
# Encode FF slides
if ff_patches.dim() == 4: # Raw images
ff_features = self.pathology_encoder(ff_patches)
else: # Pre-extracted features
ff_features = ff_patches
# Encode FFPE slides
if ffpe_patches.dim() == 4: # Raw images
ffpe_features = self.pathology_encoder(ffpe_patches)
else: # Pre-extracted features
ffpe_features = ffpe_patches
# ===== INTRA-SLIDE HYPERGRAPH PROCESSING =====
# FF slides
ff_topo_edges, ff_struct_edges = self.hypergraph_construction(
ff_features, ff_coords
)
ff_combined_edges = torch.cat([ff_topo_edges, ff_struct_edges], dim=1)
ff_agg = self.intra_slide_hypergraph(ff_features, ff_combined_edges)
# FFPE slides
ffpe_topo_edges, ffpe_struct_edges = self.hypergraph_construction(
ffpe_features, ffpe_coords
)
ffpe_combined_edges = torch.cat([ffpe_topo_edges, ffpe_struct_edges], dim=1)
ffpe_agg = self.intra_slide_hypergraph(ffpe_features, ffpe_combined_edges)
# Aggregate to patient level
ff_patient = ff_agg.mean(dim=0, keepdim=True) # (1, feature_dim)
ffpe_patient = ffpe_agg.mean(dim=0, keepdim=True) # (1, feature_dim)
# Expand to batch
ff_patient = ff_patient.expand(batch_size, -1)
ffpe_patient = ffpe_patient.expand(batch_size, -1)
# ===== INTER-SLIDE ALIGNMENT =====
# Adaptive weighting
w_ff = torch.softmax(torch.cat([
(self.slide_weight_ff.t() @ ff_patient.t()).squeeze(),
(self.slide_weight_ffpe.t() @ ffpe_patient.t()).squeeze()
]).unsqueeze(0), dim=-1)
# Cross-attention alignment using inter-slide features
inter_edges, inter_sim = self.inter_slide_hypergraph(ff_patient, ffpe_patient)
inter_features = (ff_patient + ffpe_patient) / 2 # Shared guidance
# Align FF and FFPE using inter-slide guidance
ff_aligned = ff_patient + w_ff[:, 0:1] * (
F.linear(ff_patient, self.cross_attention.W_q_p.weight) @
F.linear(inter_features, self.cross_attention.W_k_p.weight).t() /
np.sqrt(self.feature_dim)
).softmax(dim=-1) @ F.linear(inter_features, self.cross_attention.W_v_p.weight)
ffpe_aligned = ffpe_patient + w_ff[:, 1:2] * (
F.linear(ffpe_patient, self.cross_attention.W_q_p.weight) @
F.linear(inter_features, self.cross_attention.W_k_p.weight).t() /
np.sqrt(self.feature_dim)
).softmax(dim=-1) @ F.linear(inter_features, self.cross_attention.W_v_p.weight)
# Concatenate aligned representations
pathology_feat = torch.cat([ff_aligned, ffpe_aligned], dim=1) # (batch, 2*feature_dim)
pathology_feat_fused = pathology_feat[:, :self.feature_dim] # Extract for encoding
# ===== OMICS FEATURE EXTRACTION =====
if missing_modality != 'omics':
genomics_feat = self.genomics_encoder(genomics)
transcriptomics_feat = self.transcriptomics_encoder(transcriptomics)
proteomics_feat = self.proteomics_encoder(proteomics, protein_seq_embed)
# Aggregate multi-omics
omics_feat = (genomics_feat + transcriptomics_feat + proteomics_feat) / 3
else:
# Impute missing omics from memory bank
omics_feat = self.memory_bank.retrieve_missing_modality(
pathology_feat_fused, missing_type='omics', top_k=1
)
# ===== MULTIMODAL FUSION =====
if missing_modality != 'pathology':
pathology_enhanced, omics_enhanced = self.cross_attention(
pathology_feat_fused, omics_feat
)
else:
# Impute missing pathology from memory bank
pathology_enhanced = self.memory_bank.retrieve_missing_modality(
omics_feat, missing_type='pathology', top_k=1
)
omics_enhanced = omics_feat
# Concatenate fused features
fused_features = torch.cat([pathology_enhanced, omics_enhanced], dim=1)
# ===== UPDATE MEMORY BANK =====
self.memory_bank.update_prototypes(pathology_feat_fused, omics_feat)
# ===== SURVIVAL PREDICTION =====
hazard, survival = self.survival_head(fused_features)
return hazard, survival
# ============================================================================
# TRAINING UTILITIES
# ============================================================================
class SurvivalLoss(nn.Module):
"""
Negative log-likelihood loss for survival prediction.
Handles both event and censoring terms.
"""
def __init__(self):
super(SurvivalLoss, self).__init__()
def forward(self, hazard: torch.Tensor, survival: torch.Tensor,
event: torch.Tensor, time: torch.Tensor) -> torch.Tensor:
"""
Args:
hazard: (batch_size, num_time_bins)
survival: (batch_size, num_time_bins)
event: (batch_size,) binary event indicator
time: (batch_size,) observed time points
Returns:
loss: scalar loss value
"""
# Discretize time to bins
batch_size = event.shape[0]
num_bins = survival.shape[1]
time_bins = (time.float() / time.max() * (num_bins - 1)).long()
time_bins = torch.clamp(time_bins, 0, num_bins - 1)
# Event log-likelihood: log(hazard * survival)
event_ll = torch.zeros(batch_size, device=hazard.device)
for i in range(batch_size):
t = time_bins[i]
if event[i] == 1: # Event occurred
event_ll[i] = torch.log(hazard[i, t] + 1e-8)
# Censoring log-likelihood: log(survival)
cens_ll = torch.zeros(batch_size, device=survival.device)
for i in range(batch_size):
t = time_bins[i]
if event[i] == 0: # Censored
cens_ll[i] = torch.log(survival[i, t] + 1e-8)
# Combined loss
loss = -(event[event == 1].shape[0] * event_ll[event == 1].mean() +
event[event == 0].shape[0] * cens_ll[event == 0].mean() + 1e-8)
return loss
def train_m3surv(model: M3Surv, train_loader, val_loader,
num_epochs: int = 30, learning_rate: float = 1e-4,
weight_decay: float = 1e-5, device: str = 'cuda'):
"""
Training loop for M³Surv model.
Args:
model: M³Surv instance
train_loader: DataLoader for training data
val_loader: DataLoader for validation data
num_epochs: number of training epochs
learning_rate: optimization learning rate
weight_decay: L2 regularization weight
device: 'cuda' or 'cpu'
"""
model = model.to(device)
optimizer = Adam(model.parameters(), lr=learning_rate, weight_decay=weight_decay)
criterion = SurvivalLoss()
best_val_loss = float('inf')
for epoch in range(num_epochs):
# Training phase
model.train()
train_loss = 0.0
train_count = 0
for batch_idx, batch in enumerate(train_loader):
optimizer.zero_grad()
# Unpack batch
ff_patches = batch['ff_patches'].to(device)
ffpe_patches = batch['ffpe_patches'].to(device)
ff_coords = batch['ff_coords'].to(device)
ffpe_coords = batch['ffpe_coords'].to(device)
genomics = batch['genomics'].to(device)
transcriptomics = batch['transcriptomics'].to(device)
proteomics = batch['proteomics'].to(device)
protein_seq_embed = batch['protein_seq_embed'].to(device)
event = batch['event'].to(device)
time = batch['time'].to(device)
# Forward pass
hazard, survival = model(
ff_patches, ffpe_patches, ff_coords, ffpe_coords,
genomics, transcriptomics, proteomics, protein_seq_embed,
missing_modality=None
)
# Loss computation
loss = criterion(hazard, survival, event, time)
# Backward pass
loss.backward()
optimizer.step()
train_loss += loss.item()
train_count += 1
avg_train_loss = train_loss / train_count
# Validation phase
model.eval()
val_loss = 0.0
val_count = 0
with torch.no_grad():
for batch in val_loader:
ff_patches = batch['ff_patches'].to(device)
ffpe_patches = batch['ffpe_patches'].to(device)
ff_coords = batch['ff_coords'].to(device)
ffpe_coords = batch['ffpe_coords'].to(device)
genomics = batch['genomics'].to(device)
transcriptomics = batch['transcriptomics'].to(device)
proteomics = batch['proteomics'].to(device)
protein_seq_embed = batch['protein_seq_embed'].to(device)
event = batch['event'].to(device)
time = batch['time'].to(device)
hazard, survival = model(
ff_patches, ffpe_patches, ff_coords, ffpe_coords,
genomics, transcriptomics, proteomics, protein_seq_embed,
missing_modality=None
)
loss = criterion(hazard, survival, event, time)
val_loss += loss.item()
val_count += 1
avg_val_loss = val_loss / val_count
# Log progress
print(f"Epoch {epoch+1}/{num_epochs} | "
f"Train Loss: {avg_train_loss:.4f} | "
f"Val Loss: {avg_val_loss:.4f}")
# Early stopping
if avg_val_loss < best_val_loss:
best_val_loss = avg_val_loss
torch.save(model.state_dict(), 'best_m3surv_model.pth')
print(f" → Saved best model (val_loss: {best_val_loss:.4f})")
return model
def evaluate_concordance_index(model: M3Surv, test_loader, device: str = 'cuda') -> float:
"""
Evaluates model using Concordance Index (C-Index) metric.
Measures agreement between predicted risk and observed survival.
Args:
model: M³Surv instance
test_loader: DataLoader for test data
device: 'cuda' or 'cpu'
Returns:
c_index: Concordance Index score (0.5 to 1.0)
"""
from scipy.stats import rankdata
model = model.to(device)
model.eval()
all_risk_scores = []
all_times = []
all_events = []
with torch.no_grad():
for batch in test_loader:
ff_patches = batch['ff_patches'].to(device)
ffpe_patches = batch['ffpe_patches'].to(device)
ff_coords = batch['ff_coords'].to(device)
ffpe_coords = batch['ffpe_coords'].to(device)
genomics = batch['genomics'].to(device)
transcriptomics = batch['transcriptomics'].to(device)
proteomics = batch['proteomics'].to(device)
protein_seq_embed = batch['protein_seq_embed'].to(device)
hazard, survival = model(
ff_patches, ffpe_patches, ff_coords, ffpe_coords,
genomics, transcriptomics, proteomics, protein_seq_embed,
missing_modality=None
)
# Risk score = cumulative hazard
risk_scores = hazard.sum(dim=1).cpu().numpy()
all_risk_scores.extend(risk_scores)
all_times.extend(batch['time'].numpy())
all_events.extend(batch['event'].numpy())
all_risk_scores = np.array(all_risk_scores)
all_times = np.array(all_times)
all_events = np.array(all_events)
# Compute C-Index
c_index = 0.0
pair_count = 0
for i in range(len(all_times)):
for j in range(len(all_times)):
if all_events[i] == 1 and all_times[i] < all_times[j]:
pair_count += 1
if all_risk_scores[i] > all_risk_scores[j]:
c_index += 1
if pair_count > 0:
c_index = c_index / pair_count
else:
c_index = 0.5
return c_index
# ============================================================================
# EXAMPLE USAGE
# ============================================================================
if __name__ == "__main__":
print("=" * 70)
print("M³SURV: Memory-Augmented Multimodal Survival Prediction")
print("=" * 70)
# Hyperparameters
feature_dim = 768
num_time_bins = 50
num_prototypes = 10
batch_size = 4
num_epochs = 30
device = 'cuda' if torch.cuda.is_available() else 'cpu'
print(f"\n✓ Device: {device}")
# Initialize model
model = M3Surv(
feature_dim=feature_dim,
num_time_bins=num_time_bins,
num_prototypes=num_prototypes,
momentum=0.75,
spatial_threshold=100.0,
similarity_threshold=0.7
)
print(f"✓ Model initialized with {sum(p.numel() for p in model.parameters()):,} parameters")
# Create dummy data for demonstration
print("\n" + "=" * 70)
print("DEMONSTRATION WITH DUMMY DATA")
print("=" * 70)
# Simulate batch
batch = {
'ff_patches': torch.randn(batch_size, 300, feature_dim), # Pre-extracted features
'ffpe_patches': torch.randn(batch_size, 4096, feature_dim),
'ff_coords': torch.randn(batch_size, 300, 2) * 1000,
'ffpe_coords': torch.randn(batch_size, 4096, 2) * 2000,
'genomics': torch.randn(batch_size, 6),
'transcriptomics': torch.randn(batch_size, 331),
'proteomics': torch.randn(batch_size, 100),
'protein_seq_embed': torch.randn(batch_size, 100, 1280),
'event': torch.randint(0, 2, (batch_size,)).float(),
'time': torch.randint(1, 100, (batch_size,)).float()
}
# Move to device
for key in batch:
if isinstance(batch[key], torch.Tensor):
batch[key] = batch[key].to(device)
model = model.to(device)
# Forward pass
print("\n→ Forward pass with complete data...")
hazard, survival = model(
batch['ff_patches'], batch['ffpe_patches'],
batch['ff_coords'], batch['ffpe_coords'],
batch['genomics'], batch['transcriptomics'],
batch['proteomics'], batch['protein_seq_embed'],
missing_modality=None
)
print(f" Hazard shape: {hazard.shape}")
print(f" Survival shape: {survival.shape}")
print(f" Sample predictions - Hazard: {hazard[0, :5]}")
print(f" Sample predictions - Survival: {survival[0, :5]}")
# Forward pass with missing omics
print("\n→ Forward pass with MISSING OMICS (imputed from pathology)...")
hazard_missing, survival_missing = model(
batch['ff_patches'], batch['ffpe_patches'],
batch['ff_coords'], batch['ffpe_coords'],
batch['genomics'], batch['transcriptomics'],
batch['proteomics'], batch['protein_seq_embed'],
missing_modality='omics'
)
print(f" Hazard shape: {hazard_missing.shape}")
print(f" Model gracefully handled missing modality ✓")
# Forward pass with missing pathology
print("\n→ Forward pass with MISSING PATHOLOGY (imputed from omics)...")
hazard_missing_path, survival_missing_path = model(
batch['ff_patches'], batch['ffpe_patches'],
batch['ff_coords'], batch['ffpe_coords'],
batch['genomics'], batch['transcriptomics'],
batch['proteomics'], batch['protein_seq_embed'],
missing_modality='pathology'
)
print(f" Hazard shape: {hazard_missing_path.shape}")
print(f" Model gracefully handled missing modality ✓")
print("\n" + "=" * 70)
print("M³SURV implementation complete!")
print("=" * 70)Related posts, You May like to read
- 7 Shocking Truths About Knowledge Distillation: The Good, The Bad, and The Breakthrough (SAKD)
- MOSEv2: The Game-Changing Video Object Segmentation Dataset for Real-World AI Applications
- MedDINOv3: Revolutionizing Medical Image Segmentation with Adaptable Vision Foundation Models
- SurgeNetXL: Revolutionizing Surgical Computer Vision with Self-Supervised Learning
- How AI is Learning to Think Before it Segments: Understanding Seg-Zero’s Reasoning-Driven Image Analysis
- SegTrans: The Breakthrough Framework That Makes AI Segmentation Models Vulnerable to Transfer Attacks
- Universal Text-Driven Medical Image Segmentation: How MedCLIP-SAMv2 Revolutionizes Diagnostic AI
- Towards Trustworthy Breast Tumor Segmentation in Ultrasound Using AI Uncertainty
- DVIS++: The Game-Changing Decoupled Framework Revolutionizing Universal Video Segmentation
- Radar Gait Recognition Using Swin Transformers: Beyond Video Surveillance